+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM structure of yeast THO complex with Sub2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA transcription / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationnucleoplasmic THO complex / cellular response to azide / THO complex / THO complex part of transcription export complex / transcription export complex / positive regulation of transcription elongation by RNA polymerase I / Cdc73/Paf1 complex / mRNA 3'-end processing / positive regulation of transcription by RNA polymerase I / subtelomeric heterochromatin formation ...nucleoplasmic THO complex / cellular response to azide / THO complex / THO complex part of transcription export complex / transcription export complex / positive regulation of transcription elongation by RNA polymerase I / Cdc73/Paf1 complex / mRNA 3'-end processing / positive regulation of transcription by RNA polymerase I / subtelomeric heterochromatin formation / transcription-coupled nucleotide-excision repair / mRNA export from nucleus / stress granule assembly / transcription elongation by RNA polymerase II / spliceosomal complex / mRNA splicing, via spliceosome / mRNA processing / DNA recombination / nucleic acid binding / chromosome, telomeric region / molecular adaptor activity / RNA helicase activity / RNA helicase / mRNA binding / ATP hydrolysis activity / RNA binding / ATP binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Chen C / Tan M | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Bull (Beijing) / Year: 2021 Journal: Sci Bull (Beijing) / Year: 2021Title: Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex. Authors: Cong Chen / Ming Tan / Zhenfang Wu / Yuebin Zhang / Fanyang He / Yanjia Lu / Shaobai Li / Mi Cao / Guohui Li / Jian Wu / Hong Cheng / Ming Lei /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31670.map.gz emd_31670.map.gz | 135.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31670-v30.xml emd-31670-v30.xml emd-31670.xml emd-31670.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31670.png emd_31670.png | 82.8 KB | ||
| Filedesc metadata |  emd-31670.cif.gz emd-31670.cif.gz | 7.8 KB | ||
| Others |  emd_31670_additional_1.map.gz emd_31670_additional_1.map.gz | 136.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31670 http://ftp.pdbj.org/pub/emdb/structures/EMD-31670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31670 | HTTPS FTP |
-Validation report
| Summary document |  emd_31670_validation.pdf.gz emd_31670_validation.pdf.gz | 546.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31670_full_validation.pdf.gz emd_31670_full_validation.pdf.gz | 546.4 KB | Display | |
| Data in XML |  emd_31670_validation.xml.gz emd_31670_validation.xml.gz | 6.7 KB | Display | |
| Data in CIF |  emd_31670_validation.cif.gz emd_31670_validation.cif.gz | 7.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31670 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31670 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31670 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31670 | HTTPS FTP |
-Related structure data
| Related structure data |  7v2yMC  7v2wC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_31670.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31670.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
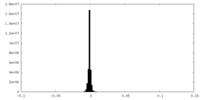
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_31670_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
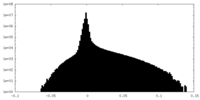
| Density Histograms |
- Sample components
Sample components
-Entire : THO-Sub2 Complex
| Entire | Name: THO-Sub2 Complex |
|---|---|
| Components |
|
-Supramolecule #1: THO-Sub2 Complex
| Supramolecule | Name: THO-Sub2 Complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: yeast THO
| Supramolecule | Name: yeast THO / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Sub2
| Supramolecule | Name: Sub2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #6 |
|---|
-Macromolecule #1: THO complex subunit HPR1
| Macromolecule | Name: THO complex subunit HPR1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87.938547 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSNTEELIQN SIGFLQKTFK ALPVSFDSIR HEPLPSSMLH ASVLNFEWEP LEKNISAIHD RDSLIDIILK RFIIDSMTNA IEDEEENNL EKGLLNSCIG LDFVYNSRFN RSNPASWGNT FFELFSTIID LLNSPSTFLK FWPYAESRIE WFKMNTSVEP V SLGESNLI ...String: MSNTEELIQN SIGFLQKTFK ALPVSFDSIR HEPLPSSMLH ASVLNFEWEP LEKNISAIHD RDSLIDIILK RFIIDSMTNA IEDEEENNL EKGLLNSCIG LDFVYNSRFN RSNPASWGNT FFELFSTIID LLNSPSTFLK FWPYAESRIE WFKMNTSVEP V SLGESNLI SYKQPLYEKL RHWNDILAKL ENNDILNTVK HYNMKYKLEN FLSELLPINE ESNFNRSASI SALQESDNEW NR SARERES NRSSDVIFAA DYNFVFYHLI ICPIEFAFSD LEYKNDVDRS LSPLLDAILE IEENFYSKIK MNNRTRYSLE EAL NTEYYA NYDVMTPKLP VYMKHSNAMK MDRNEFWANL QNIKESDDYT LRPTIMDISL SNTTCLYKQL TQEDDDYYRK QFIL QLCFT TNLIRNLISS DETRNFYKSC YLRENPLSDI DFENLDEVNK KRGLNLCSYI CDNRVLKFYK IKDPDFYRVI RKLMS SDEK FTTAKIDGFK EFQNFRISKE KIPPPAFDET FKKFTFIKMG NKLINNVWKI PTGLDKIEQE VKKPEGVYEA AQAKWE SKI SSETSGGEAK DEIIRQWQTL RFLRSRYLFD FDKVNEKTGV DGLFEEPRKV EALDDSFKEK LLYKINQEHR KKLQDAR EY KIGKERKKRA LEEEASFPER EQKIKSQRIN SASQTEGDEL KSEQTQPKGE ISEENTKIKS SEVSSQDPDS GVAGEFAP Q NTTAQLENPK TEDNNAATSN ISNGSSTQDM K UniProtKB: THO complex subunit HPR1 |
-Macromolecule #2: THO complex subunit 2
| Macromolecule | Name: THO complex subunit 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 184.163734 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAEQTLLSKL NALSQKVIPP ASPSQASILT EEVIRNWPER SKTLCSDFTA LESNDEKEDW LRTLFIELFD FINKNDENSP LKLSDVASF TNELVNHERQ VSQASIVGKM FIAVSSTVPN INDLTTISLC KLIPSLHEEL FKFSWISSKL LNKEQTTLLR H LLKKSKYE ...String: MAEQTLLSKL NALSQKVIPP ASPSQASILT EEVIRNWPER SKTLCSDFTA LESNDEKEDW LRTLFIELFD FINKNDENSP LKLSDVASF TNELVNHERQ VSQASIVGKM FIAVSSTVPN INDLTTISLC KLIPSLHEEL FKFSWISSKL LNKEQTTLLR H LLKKSKYE LKKYNLLVEN SVGYGQLVAL LILAYYDPDN FSKVSAYLKE IYHIMGKYSL DSIRTLDVIL NVSSQFITEG YK FFIALLR KSDSWPSSHV ANNSNYSSLN EGGNMIAANI ISFNLSQYNE EVDKENYERY MDMCCILLKN GFVNFYSIWD NVK PEMEFL QEYIQNLETE LEEESTKGVE NPLAMAAALS TENETDEDNA LVVNDDVNMK DKISEETNAD IESKGKQKTQ QDIL LFGKI KLLERLLIHG CVIPVIHVLK QYPKVLYVSE SLSRYLGRVF EYLLNPLYTS MTSSGESKDM ATALMITRID NGILA HKPR LIHKYKTHEP FESLELNSSY VFYYSEWNSN LTPFASVNDL FENSHIYLSI IGPYLGRIPT LLSKISRIGV ADIQKN HGS ESLHVTIDKW IDYVRKFIFP ATSLLQNNPI ATSEVYELMK FFPFEKRYFI YNEMMTKLSQ DILPLKVSFN KAEREAK SI LKALSIDTIA KESRRFAKLI STNPLASLVP AVKQIENYDK VSELVVYTTK YFNDFAYDVL QFVLLLRLTY NRPAVQFD G VNQAMWVQRL SIFIAGLAKN CPNMDISNII TYILKTLHNG NIIAVSILKE LIITVGGIRD LNEVNMKQLL MLNSGSPLK QYARHLIYDF RDDNSVISSR LTSFFTDQSA ISEIILLLYT LNLKANTQNS HYKILSTRCD EMNTLLWSFI ELIKHCLKGK AFEENVLPF VELNNRFHLS TPWTFHIWRD YLDNQLNSNE NFSIDELIEG AEFSDVDLTK ISKDLFTTFW RLSLYDIHFD K SLYDERKN ALSGENTGHM SNRKKHLIQN QIKDILVTGI SHQRAFKKTS EFISEKSNVW NKDCGEDQIK IFLQNCVVPR VL FSPSDAL FSSFFIFMAF RTENLMSILN TCITSNILKT LLFCCTSSEA GNLGLFFTDV LKKLEKMRLN GDFNDQASRK LYE WHSVIT EQVIDLLSEK NYMSIRNGIE FMKHVTSVFP VVKAHIQLVY TTLEENLINE EREDIKLPSS ALIGHLKARL KDAL ELDEF CTLTEEEAEQ KRIREMELEE IKNYETACQN EQKQVALRKQ LELNKSQRLQ NDPPKSVASG SAGLNSKDRY TYSRN EPVI PTKPSSSQWS YSKVTRHVDD INHYLATNHL QKAISLVEND DETRNLRKLS KQNMPIFDFR NSTLEIFERY FRTLIQ NPQ NPDFAEKIDS LKRYIKNISR EPYPDTTSSY SEAAAPEYTK RSSRYSGNAG GKDGYGSSNY RGPSNDRSAP KNIKPIS SY AHKRSELPTR PSKSKTYNDR SRALRPTGPD RGDGFDQRDN RLREEYKKNS SQRSQLRFPE KPFQEGKDSS KANPYQAS S YKRDSPSENE EKPNKRFKKD ETIRNKFQTQ DYRNTRDSGA AHRANENQRY NGNRKSNTQA LPQGPKGGNY VSRYQR UniProtKB: THO complex subunit 2 |
-Macromolecule #3: Protein TEX1
| Macromolecule | Name: Protein TEX1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 47.229652 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTIGAVDIL NQKTITSEVA ASVTSKYLQS TFSKGNTSHI EDKRFIHVSS RSHSRFTSTP ITPNEILSLK FHVSGSSMAY SRMDGSLTV WFIKDASFDK SVEVYIPDCC GSDKLATDLS WNPTSLNQIA VVSNSSEISL LLINEKSLTA SKLRTLSLGS K TKVNTCLY ...String: MSTIGAVDIL NQKTITSEVA ASVTSKYLQS TFSKGNTSHI EDKRFIHVSS RSHSRFTSTP ITPNEILSLK FHVSGSSMAY SRMDGSLTV WFIKDASFDK SVEVYIPDCC GSDKLATDLS WNPTSLNQIA VVSNSSEISL LLINEKSLTA SKLRTLSLGS K TKVNTCLY DPLGNWLLAA TKSEKIYLFD VKKDHSSVCS LNISDISQED NDVVYSLAWS NGGSHIFIGF KSGYLAILKA KH GILEVCT KIKAHTGPIT EIKMDPWGRN FITGSIDGNC YVWNMKSLCC ELIINDLNSA VTTLDVCHLG KILGICTEDE MVY FYDLNS GNLLHSKSLA NYKTDPVLKF YPDKSWYIMS GKNDTLSNHF VKNEKNLITY WKDMFDNTMI EKRRKNNGGG NNHN KRTSK NTDRIGKDRP SRFNSKK UniProtKB: Protein TEX1 |
-Macromolecule #4: THO complex subunit MFT1
| Macromolecule | Name: THO complex subunit MFT1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.055012 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPLSQKQIDQ VRTKVHYSEV DTPFNKYLDI LGKVTKLTGS IINGTLSNDD SKIEKLTEQN ISQLKESAHL RFLDLQSSID TKKVADENW ETCQQETLAK LENLKDKLPD IKSIHSKLLL RIGKLQGLYD SVQVINREVE GLSEGRTSLV VTRAEWEKEL G TDLVKFLI ...String: MPLSQKQIDQ VRTKVHYSEV DTPFNKYLDI LGKVTKLTGS IINGTLSNDD SKIEKLTEQN ISQLKESAHL RFLDLQSSID TKKVADENW ETCQQETLAK LENLKDKLPD IKSIHSKLLL RIGKLQGLYD SVQVINREVE GLSEGRTSLV VTRAEWEKEL G TDLVKFLI EKNYLKLVDP GLKKDSSEER YRIYDDFSKG PKELESINAS MKSDIENVRQ EVSSYKEKWL RDAEIFGKIT SI FKEELLK RDGLLNEAEG DNIDEDYESD EDEERKERFK RQRSMVEVNT IENVDEKEES DHEYDDQEDE ENEEEDDMEV DVE DIKEDN EVDGESSQQE DNSRQGNNEE TDKETGVIEE PDAVNDAEEA DSDHSSRKLG GTTSDFSASS SVEEVK UniProtKB: THO complex subunit MFT1 |
-Macromolecule #5: THO complex subunit THP2
| Macromolecule | Name: THO complex subunit THP2 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.340264 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTKEEGRTYF ESLCEEEQSL QESQTHLLNI LDILSVLADP RSSDDLLTES LKKLPDLHRE LINSSIRLRY DKYQTREAQL LEDTKTGRD VAAGVQNPKS ISEYYSTFEH LNRDTLRYIN LLKRLSVDLA KQVEVSDPSV TVYEMDKWVP SEKLQGILEQ Y CAPDTDIR ...String: MTKEEGRTYF ESLCEEEQSL QESQTHLLNI LDILSVLADP RSSDDLLTES LKKLPDLHRE LINSSIRLRY DKYQTREAQL LEDTKTGRD VAAGVQNPKS ISEYYSTFEH LNRDTLRYIN LLKRLSVDLA KQVEVSDPSV TVYEMDKWVP SEKLQGILEQ Y CAPDTDIR GVDAQIKNYL DQIKMARAKF GLENKYSLKE RLSTLTKELN HWRKEWDDIE MLMFGDDAHS MKKMIQKIDS LK SEINAPS ESYPVDKEGD IVLE UniProtKB: THO complex subunit THP2 |
-Macromolecule #6: ATP-dependent RNA helicase SUB2
| Macromolecule | Name: ATP-dependent RNA helicase SUB2 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA helicase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50.375859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSHEGEEDLL EYSDNEQEIQ IDASKAAEAG ETGAATSATE GDNNNNTAAG DKKGSYVGIH STGFKDFLLK PELSRAIIDC GFEHPSEVQ QHTIPQSIHG TDVLCQAKSG LGKTAVFVLS TLQQLDPVPG EVAVVVICNA RELAYQIRNE YLRFSKYMPD V KTAVFYGG ...String: MSHEGEEDLL EYSDNEQEIQ IDASKAAEAG ETGAATSATE GDNNNNTAAG DKKGSYVGIH STGFKDFLLK PELSRAIIDC GFEHPSEVQ QHTIPQSIHG TDVLCQAKSG LGKTAVFVLS TLQQLDPVPG EVAVVVICNA RELAYQIRNE YLRFSKYMPD V KTAVFYGG TPISKDAELL KNKDTAPHIV VATPGRLKAL VREKYIDLSH VKNFVIDECD KVLEELDMRR DVQEIFRATP RD KQVMMFS ATLSQEIRPI CRRFLQNPLE IFVDDEAKLT LHGLQQYYIK LEEREKNRKL AQLLDDLEFN QVIIFVKSTT RAN ELTKLL NASNFPAITV HGHMKQEERI ARYKAFKDFE KRICVSTDVF GRGIDIERIN LAINYDLTNE ADQYLHRVGR AGRF GTKGL AISFVSSKED EEVLAKIQER FDVKIAEFPE EGIDPSTYLN N UniProtKB: ATP-dependent RNA helicase SUB2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 284361 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)