+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30370 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of E.coli MlaFEB | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mla complex / Lipid transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationATPase-coupled transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex / ATP binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Zhou C / Shi H | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2021 Journal: J Mol Biol / Year: 2021Title: Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa. Authors: Changping Zhou / Huigang Shi / Manfeng Zhang / Lijun Zhou / Le Xiao / Shasha Feng / Wonpil Im / Min Zhou / Xinzheng Zhang / Yihua Huang /   Abstract: The outer membrane (OM) of Gram-negative bacteria, which consists of lipopolysaccharides (LPS) in the outer leaflet and phospholipids (PLs) in the inner leaflet, plays a key role in antibiotic ...The outer membrane (OM) of Gram-negative bacteria, which consists of lipopolysaccharides (LPS) in the outer leaflet and phospholipids (PLs) in the inner leaflet, plays a key role in antibiotic resistance and pathogen virulence. The maintenance of lipid asymmetry (Mla) pathway is known to be involved in PL transport and contributes to the lipid homeostasis of the OM, yet the underlying molecular mechanism and the directionality of PL transport in this pathway remain elusive. Here, we reported the cryo-EM structures of the ATP-binding cassette (ABC) transporter MlaFEBD from P. areuginosa, the core complex in the Mla pathway, in nucleotide-free (apo)-, ADP (ATP + vanadate)- and ATP (AMPPNP)-bound states as well as the structures of MlaFEB from E. coli in apo- and AMPPNP-bound states at a resolution range of 3.4-3.9 Å. The structures show that the MlaFEBD complex contains a total of twelve protein molecules with a stoichiometry of MlaFEBD, and binds a plethora of PLs at different locations. In contrast to canonical ABC transporters, nucleotide binding fails to trigger significant conformational changes of both MlaFEBD and MlaFEB in the nucleotide-binding and transmembrane domains of the ABC transporter, correlated with their low ATPase activities exhibited in both detergent micelles and lipid nanodiscs. Intriguingly, PLs or detergents appeared to relocate to the membrane-proximal end from the distal end of the hydrophobic tunnel formed by the MlaD hexamer in MlaFEBD upon addition of ATP, indicating that retrograde PL transport might occur in the tunnel in an ATP-dependent manner. Site-specific photocrosslinking experiment confirms that the substrate-binding pocket in the dimeric MlaE and the MlaD hexamer are able to bind PLs in vitro, in line with the notion that MlaFEBD complex functions as a PL transporter. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30370.map.gz emd_30370.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30370-v30.xml emd-30370-v30.xml emd-30370.xml emd-30370.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
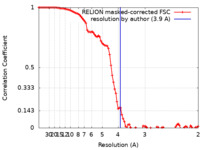
| FSC (resolution estimation) |  emd_30370_fsc.xml emd_30370_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_30370.png emd_30370.png | 53.7 KB | ||
| Filedesc metadata |  emd-30370.cif.gz emd-30370.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30370 http://ftp.pdbj.org/pub/emdb/structures/EMD-30370 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30370 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30370 | HTTPS FTP |
-Validation report
| Summary document |  emd_30370_validation.pdf.gz emd_30370_validation.pdf.gz | 580.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30370_full_validation.pdf.gz emd_30370_full_validation.pdf.gz | 580.4 KB | Display | |
| Data in XML |  emd_30370_validation.xml.gz emd_30370_validation.xml.gz | 9.9 KB | Display | |
| Data in CIF |  emd_30370_validation.cif.gz emd_30370_validation.cif.gz | 13.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30370 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30370 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30370 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30370 | HTTPS FTP |
-Related structure data
| Related structure data |  7ch7MC  7ch6C  7ch8C  7ch9C  7chaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30370.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30370.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : E.coli Mla FEB complex
| Entire | Name: E.coli Mla FEB complex |
|---|---|
| Components |
|
-Supramolecule #1: E.coli Mla FEB complex
| Supramolecule | Name: E.coli Mla FEB complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Lipid asymmetry maintenance ABC transporter permease subunit MlaE
| Macromolecule | Name: Lipid asymmetry maintenance ABC transporter permease subunit MlaE type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.885162 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLLNALASLG HKGIKTLRTF GRAGLMLFNA LVGKPEFRKH APLLVRQLYN VGVLSMLIIV VSGVFIGMVL GLQGYLVLTT YSAETSLGM LVALSLLREL GPVVAALLFA GRAGSALTAE IGLMRATEQL SSMEMMAVDP LRRVISPRFW AGVISLPLLT V IFVAVGIW ...String: MLLNALASLG HKGIKTLRTF GRAGLMLFNA LVGKPEFRKH APLLVRQLYN VGVLSMLIIV VSGVFIGMVL GLQGYLVLTT YSAETSLGM LVALSLLREL GPVVAALLFA GRAGSALTAE IGLMRATEQL SSMEMMAVDP LRRVISPRFW AGVISLPLLT V IFVAVGIW GGSLVGVSWK GIDSGFFWSA MQNAVDWRMD LVNCLIKSVV FAITVTWISL FNGYDAIPTS AGISRATTRT VV HSSLAVL GLDFVLTALM FGN UniProtKB: Intermembrane phospholipid transport system permease protein MlaE |
-Macromolecule #2: Phospholipid ABC transporter ATP-binding protein MlaF
| Macromolecule | Name: Phospholipid ABC transporter ATP-binding protein MlaF / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.128801 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEQSVANLVD MRDVSFTRGN RCIFDNISLT VPRGKITAIM GPSGIGKTTL LRLIGGQIAP DHGEILFDGE NIPAMSRSRL YTVRKRMSM LFQSGALFTD MNVFDNVAYP LREHTQLPAP LLHSTVMMKL EAVGLRGAAK LMPSELSGGM ARRAALARAI A LEPDLIMF ...String: MEQSVANLVD MRDVSFTRGN RCIFDNISLT VPRGKITAIM GPSGIGKTTL LRLIGGQIAP DHGEILFDGE NIPAMSRSRL YTVRKRMSM LFQSGALFTD MNVFDNVAYP LREHTQLPAP LLHSTVMMKL EAVGLRGAAK LMPSELSGGM ARRAALARAI A LEPDLIMF DEPFVGQDPI TMGVLVKLIS ELNSALGVTC VVVSHDVPEV LSIADHAWIL ADKKIVAHGS AQALQANPDP RV RQFLDGI ADGPVPFRYP AGDYHADLLP GS UniProtKB: Phospholipid ABC transporter ATP-binding protein MlaF |
-Macromolecule #3: Lipid asymmetry maintenance protein MlaB
| Macromolecule | Name: Lipid asymmetry maintenance protein MlaB / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.690313 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSESLSWMQT GDTLALSGEL DQDVLLPLWE MREEAVKGIT CIDLSRVSRV DTGGLALLLH LIDLAKKQGN NVTLQGVNDK VYTLAKLYN LPADVLPR UniProtKB: Lipid asymmetry maintenance protein MlaB |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: DIFFRACTION / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)