[English] 日本語
 Yorodumi
Yorodumi- EMDB-28860: Top-down design of protein architectures with reinforcement learning -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Top-down design of protein architectures with reinforcement learning | |||||||||
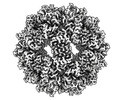
 Map data Map data | Sharpened Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nanoparticle / capsid / oligomer / de novo design / rosetta / DE NOVO PROTEIN / reinforcement learning | |||||||||
| Biological species |  | |||||||||
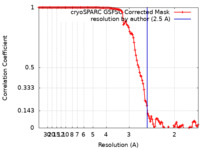
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Borst AJ / Baker D | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Top-down design of protein architectures with reinforcement learning. Authors: Isaac D Lutz / Shunzhi Wang / Christoffer Norn / Alexis Courbet / Andrew J Borst / Yan Ting Zhao / Annie Dosey / Longxing Cao / Jinwei Xu / Elizabeth M Leaf / Catherine Treichel / Patrisia ...Authors: Isaac D Lutz / Shunzhi Wang / Christoffer Norn / Alexis Courbet / Andrew J Borst / Yan Ting Zhao / Annie Dosey / Longxing Cao / Jinwei Xu / Elizabeth M Leaf / Catherine Treichel / Patrisia Litvicov / Zhe Li / Alexander D Goodson / Paula Rivera-Sánchez / Ana-Maria Bratovianu / Minkyung Baek / Neil P King / Hannele Ruohola-Baker / David Baker /     Abstract: As a result of evolutionary selection, the subunits of naturally occurring protein assemblies often fit together with substantial shape complementarity to generate architectures optimal for function ...As a result of evolutionary selection, the subunits of naturally occurring protein assemblies often fit together with substantial shape complementarity to generate architectures optimal for function in a manner not achievable by current design approaches. We describe a "top-down" reinforcement learning-based design approach that solves this problem using Monte Carlo tree search to sample protein conformers in the context of an overall architecture and specified functional constraints. Cryo-electron microscopy structures of the designed disk-shaped nanopores and ultracompact icosahedra are very close to the computational models. The icosohedra enable very-high-density display of immunogens and signaling molecules, which potentiates vaccine response and angiogenesis induction. Our approach enables the top-down design of complex protein nanomaterials with desired system properties and demonstrates the power of reinforcement learning in protein design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28860.map.gz emd_28860.map.gz | 167.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28860-v30.xml emd-28860-v30.xml emd-28860.xml emd-28860.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28860_fsc.xml emd_28860_fsc.xml | 11.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_28860.png emd_28860.png | 177.2 KB | ||
| Filedesc metadata |  emd-28860.cif.gz emd-28860.cif.gz | 5.4 KB | ||
| Others |  emd_28860_additional_1.map.gz emd_28860_additional_1.map.gz emd_28860_half_map_1.map.gz emd_28860_half_map_1.map.gz emd_28860_half_map_2.map.gz emd_28860_half_map_2.map.gz | 85.5 MB 164.5 MB 164.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28860 http://ftp.pdbj.org/pub/emdb/structures/EMD-28860 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28860 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28860 | HTTPS FTP |
-Related structure data
| Related structure data |  8f54MC  8f4xC  8f53C M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28860.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28860.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
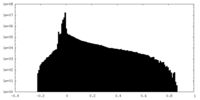
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened Map
| File | emd_28860_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
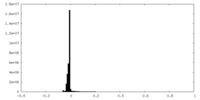
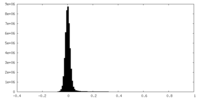
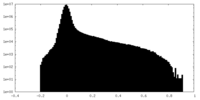
| Density Histograms |
-Half map: Half Map B
| File | emd_28860_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
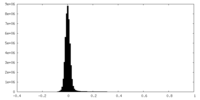
| Density Histograms |
-Half map: Half Map A
| File | emd_28860_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
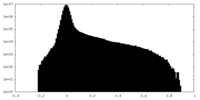
| Density Histograms |
- Sample components
Sample components
-Entire : RC_I_1
| Entire | Name: RC_I_1 |
|---|---|
| Components |
|
-Supramolecule #1: RC_I_1
| Supramolecule | Name: RC_I_1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: RC_I_1
| Macromolecule | Name: RC_I_1 / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.828097 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: PDEDLKAELA ATEAIWLLRQ GRPEEVWKLM QRLYEKGDPA LWAVLRALLR SGDEIAILIA WNFMQRI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/2 / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Average electron dose: 61.155 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)