[English] 日本語
 Yorodumi
Yorodumi- EMDB-28544: Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like envi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane protein / SARS-CoV / SARS-CoV-2 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated activation of host inflammasome-mediated signal transduction / Maturation of protein 3a / Defective ABCA1 causes TGD / high-density lipoprotein particle receptor binding / peptidyl-methionine modification / HDL clearance / spherical high-density lipoprotein particle / Scavenging by Class B Receptors / negative regulation of response to cytokine stimulus / protein oxidation ...symbiont-mediated activation of host inflammasome-mediated signal transduction / Maturation of protein 3a / Defective ABCA1 causes TGD / high-density lipoprotein particle receptor binding / peptidyl-methionine modification / HDL clearance / spherical high-density lipoprotein particle / Scavenging by Class B Receptors / negative regulation of response to cytokine stimulus / protein oxidation / regulation of intestinal cholesterol absorption / vitamin transport / blood vessel endothelial cell migration / cholesterol import / negative regulation of heterotypic cell-cell adhesion / apolipoprotein receptor binding / apolipoprotein A-I receptor binding / ABC transporters in lipid homeostasis / negative regulation of cell adhesion molecule production / Microbial modulation of RIPK1-mediated regulated necrosis / negative regulation of cytokine production involved in immune response / HDL assembly / high-density lipoprotein particle binding / negative regulation of very-low-density lipoprotein particle remodeling / glucocorticoid metabolic process / phosphatidylcholine biosynthetic process / acylglycerol homeostasis / Translation of Structural Proteins / Virion Assembly and Release / phosphatidylcholine-sterol O-acyltransferase activator activity / positive regulation of phospholipid efflux / Chylomicron remodeling / SARS-CoV-1-mediated effects on programmed cell death / cellular response to lipoprotein particle stimulus / Chylomicron assembly / high-density lipoprotein particle clearance / phospholipid efflux / chylomicron / high-density lipoprotein particle remodeling / reverse cholesterol transport / positive regulation of cholesterol metabolic process / lipid storage / high-density lipoprotein particle assembly / phospholipid homeostasis / chemorepellent activity / low-density lipoprotein particle / lipoprotein biosynthetic process / cholesterol transfer activity / cholesterol transport / high-density lipoprotein particle / very-low-density lipoprotein particle / endothelial cell proliferation / regulation of Cdc42 protein signal transduction / HDL remodeling / : / cholesterol efflux / adrenal gland development / Scavenging by Class A Receptors / triglyceride homeostasis / voltage-gated calcium channel complex / negative chemotaxis / negative regulation of interleukin-1 beta production / cholesterol binding / cholesterol biosynthetic process / amyloid-beta formation / positive regulation of Rho protein signal transduction / host cell Golgi membrane / positive regulation of cholesterol efflux / monoatomic ion channel activity / endocytic vesicle / negative regulation of tumor necrosis factor-mediated signaling pathway / Scavenging of heme from plasma / Retinoid metabolism and transport / cholesterol metabolic process / SARS-CoV-1 targets host intracellular signalling and regulatory pathways / positive regulation of stress fiber assembly / Attachment and Entry / heat shock protein binding / voltage-gated potassium channel complex / endocytic vesicle lumen / positive regulation of substrate adhesion-dependent cell spreading / positive regulation of phagocytosis / cholesterol homeostasis / integrin-mediated signaling pathway / Post-translational protein phosphorylation / Heme signaling / PPARA activates gene expression / phospholipid binding / : / negative regulation of inflammatory response / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / SARS-CoV-1 activates/modulates innate immune responses / Platelet degranulation / extracellular vesicle / amyloid-beta binding / cytoplasmic vesicle / secretory granule lumen / blood microparticle / early endosome / protein stabilization Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  Severe acute respiratory syndrome coronavirus Severe acute respiratory syndrome coronavirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Miller AN / Houlihan PR / Matamala E / Cabezas-Bratesco D / Lee GY / Cristofori-Armstrong B / Dilan TL / Sanchez-Martinez S / Matthies D / Yan R ...Miller AN / Houlihan PR / Matamala E / Cabezas-Bratesco D / Lee GY / Cristofori-Armstrong B / Dilan TL / Sanchez-Martinez S / Matthies D / Yan R / Yu Z / Ren D / Brauchi SE / Clapham DE | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation | Journal: bioRxiv / Year: 2022 Title: The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins. Authors: Alexandria N Miller / Patrick R Houlihan / Ella Matamala / Deny Cabezas-Bratesco / Gi Young Lee / Ben Cristofori-Armstrong / Tanya L Dilan / Silvia Sanchez-Martinez / Doreen Matthies / Rui ...Authors: Alexandria N Miller / Patrick R Houlihan / Ella Matamala / Deny Cabezas-Bratesco / Gi Young Lee / Ben Cristofori-Armstrong / Tanya L Dilan / Silvia Sanchez-Martinez / Doreen Matthies / Rui Yan / Zhiheng Yu / Dejian Ren / Sebastian E Brauchi / David E Clapham Abstract: The severe acute respiratory syndrome associated coronavirus 2 (SARS-CoV-2) and SARS-CoV-1 accessory protein Orf3a colocalizes with markers of the plasma membrane, endocytic pathway, and Golgi ...The severe acute respiratory syndrome associated coronavirus 2 (SARS-CoV-2) and SARS-CoV-1 accessory protein Orf3a colocalizes with markers of the plasma membrane, endocytic pathway, and Golgi apparatus. Some reports have led to annotation of both Orf3a proteins as a viroporin. Here we show that neither SARS-CoV-2 nor SARS-CoV-1 form functional ion conducting pores and that the conductances measured are common contaminants in overexpression and with high levels of protein in reconstitution studies. Cryo-EM structures of both SARS-CoV-2 and SARS-CoV-1 Orf3a display a narrow constriction and the presence of a basic aqueous vestibule, which would not favor cation permeation. We observe enrichment of the late endosomal marker Rab7 upon SARS-CoV-2 Orf3a overexpression, and co-immunoprecipitation with VPS39. Interestingly, SARS-CoV-1 Orf3a does not cause the same cellular phenotype as SARS-CoV-2 Orf3a and does not interact with VPS39. To explain this difference, we find that a divergent, unstructured loop of SARS-CoV-2 Orf3a facilitates its binding with VPS39, a HOPS complex tethering protein involved in late endosome and autophagosome fusion with lysosomes. We suggest that the added loop enhances SARS-CoV-2 Orf3a ability to co-opt host cellular trafficking mechanisms for viral exit or host immune evasion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28544.map.gz emd_28544.map.gz | 4.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28544-v30.xml emd-28544-v30.xml emd-28544.xml emd-28544.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_28544.png emd_28544.png | 108.3 KB | ||
| Filedesc metadata |  emd-28544.cif.gz emd-28544.cif.gz | 6.2 KB | ||
| Others |  emd_28544_half_map_1.map.gz emd_28544_half_map_1.map.gz emd_28544_half_map_2.map.gz emd_28544_half_map_2.map.gz | 48.5 MB 48.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28544 http://ftp.pdbj.org/pub/emdb/structures/EMD-28544 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28544 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28544 | HTTPS FTP |
-Related structure data
| Related structure data |  8eqsMC  8eqjC  8eqtC  8equC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28544.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28544.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.844 Å | ||||||||||||||||||||||||||||||||||||
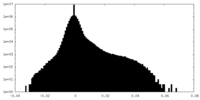
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28544_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
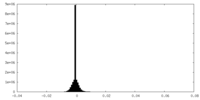
| Density Histograms |
-Half map: #1
| File | emd_28544_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
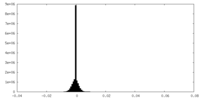
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like envi...
| Entire | Name: Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc |
|---|---|
| Components |
|
-Supramolecule #1: Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like envi...
| Supramolecule | Name: Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ORF3a protein
| Macromolecule | Name: ORF3a protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Severe acute respiratory syndrome coronavirus Severe acute respiratory syndrome coronavirus |
| Molecular weight | Theoretical: 36.268219 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDLFMRFFTL GSITAQPVKI DNASPASTVH ATATIPLQAS LPFGWLVIGV AFLAVFQSAT KIIALNKRWQ LALYKGFQFI CNLLLLFVT IYSHLLLVAA GMEAQFLYLY ALIYFLQCIN ACRIIMRCWL CWKCKSKNPL LYDANYFVCW HTHNYDYCIP Y NSVTDTIV ...String: MDLFMRFFTL GSITAQPVKI DNASPASTVH ATATIPLQAS LPFGWLVIGV AFLAVFQSAT KIIALNKRWQ LALYKGFQFI CNLLLLFVT IYSHLLLVAA GMEAQFLYLY ALIYFLQCIN ACRIIMRCWL CWKCKSKNPL LYDANYFVCW HTHNYDYCIP Y NSVTDTIV VTEGDGISTP KLKEDYQIGG YSEDRHSGVK DYVVVHGYFT EVYYQLESTQ ITTDTGIENA TFFIFNKLVK DP PNVQIHT IDGSSGVANP AMDPIYDEPT TTTSVPLGGR GLEVLFQGPG SGQLVGSGGL EGGGGWSHPQ FEKGGGSGGG SGG GSWSHP QFEK UniProtKB: ORF3a protein |
-Macromolecule #2: Apolipoprotein A-I
| Macromolecule | Name: Apolipoprotein A-I / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.704729 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GHHHHHHHDY DIPTTENLYF QGSTFSKLRE QLGPVTQEFW DNLEKETEGL RQEMSKDLEE VKAKVQPYLD DFQKKWQEEM ELYRQKVEP LRAELQEGAR QKLHELQEKL SPLGEEMRDR ARAHVDALRT HLAPYSDELR QRLAARLEAL KENGGARLAE Y HAKATEHL ...String: GHHHHHHHDY DIPTTENLYF QGSTFSKLRE QLGPVTQEFW DNLEKETEGL RQEMSKDLEE VKAKVQPYLD DFQKKWQEEM ELYRQKVEP LRAELQEGAR QKLHELQEKL SPLGEEMRDR ARAHVDALRT HLAPYSDELR QRLAARLEAL KENGGARLAE Y HAKATEHL STLSEKAKPA LEDLRQGLLP VLESFKVSFL SALEEYTKKL NTQ UniProtKB: Apolipoprotein A-I |
-Macromolecule #3: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
| Macromolecule | Name: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine / type: ligand / ID: 3 / Number of copies: 2 / Formula: PEE |
|---|---|
| Molecular weight | Theoretical: 744.034 Da |
| Chemical component information |  ChemComp-PEE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 13970 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 162607 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 3.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION (ver. 3.1) |
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8eqs: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)