[English] 日本語
 Yorodumi
Yorodumi- EMDB-28147: Rabbit muscle aldolase determined using single-particle cryo-EM w... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera. | ||||||||||||
 Map data Map data | sharpened and masked map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | glycolysis / SUGAR BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of Arp2/3 complex-mediated actin nucleation / fructose-bisphosphate aldolase / fructose-bisphosphate aldolase activity / M band / I band / glycolytic process / protein homotetramerization / positive regulation of cell migration Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
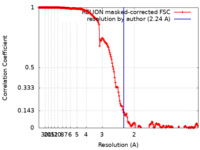
| Method | single particle reconstruction / cryo EM / Resolution: 2.24 Å | ||||||||||||
 Authors Authors | Peng R / Fu X / Mendez JH / Randolph PH / Bammes B / Stagg SM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: J Struct Biol X / Year: 2023 Journal: J Struct Biol X / Year: 2023Title: Characterizing the resolution and throughput of the Apollo direct electron detector. Authors: Ruizhi Peng / Xiaofeng Fu / Joshua H Mendez / Peter S Randolph / Benjamin E Bammes / Scott M Stagg /  Abstract: Advances in electron detection have been essential to the success of high-resolution cryo-EM structure determination. A new generation of direct electron detector called the Apollo, has been ...Advances in electron detection have been essential to the success of high-resolution cryo-EM structure determination. A new generation of direct electron detector called the Apollo, has been developed by Direct Electron. The Apollo uses a novel event-based MAPS detector custom designed for ultra-fast electron counting. We have evaluated this new camera, finding that it delivers high detective quantum efficiency (DQE) and low coincidence loss, enabling high-quality electron counting data acquisition at up to nearly 80 input electrons per pixel per second. We further characterized the performance of Apollo for single particle cryo-EM on real biological samples. Using mouse apoferritin, Apollo yielded better than 1.9 Å resolution reconstructions at all three tested dose rates from a half-day data collection session each. With longer collection time and improved specimen preparation, mouse apoferritin was reconstructed to 1.66 Å resolution. Applied to a more challenging small protein aldolase, we obtained a 2.24 Å resolution reconstruction. The high quality of the map indicates that the Apollo has sufficiently high DQE to reconstruct smaller proteins and complexes with high-fidelity. Our results demonstrate that the Apollo camera performs well across a broad range of dose rates and is capable of capturing high quality data that produce high-resolution reconstructions for large and small single particle samples. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28147.map.gz emd_28147.map.gz | 34.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28147-v30.xml emd-28147-v30.xml emd-28147.xml emd-28147.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28147_fsc.xml emd_28147_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_28147.png emd_28147.png | 137.7 KB | ||
| Masks |  emd_28147_msk_1.map emd_28147_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-28147.cif.gz emd-28147.cif.gz | 6.1 KB | ||
| Others |  emd_28147_half_map_1.map.gz emd_28147_half_map_1.map.gz emd_28147_half_map_2.map.gz emd_28147_half_map_2.map.gz | 474.2 MB 474.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28147 http://ftp.pdbj.org/pub/emdb/structures/EMD-28147 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28147 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28147 | HTTPS FTP |
-Related structure data
| Related structure data |  8ehgMC  8emqC  8en7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_28147.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28147.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened and masked map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.599 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_28147_msk_1.map emd_28147_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_28147_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_28147_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Aldolase from rabbit muscle
| Entire | Name: Aldolase from rabbit muscle |
|---|---|
| Components |
|
-Supramolecule #1: Aldolase from rabbit muscle
| Supramolecule | Name: Aldolase from rabbit muscle / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Plays a key role in glycolysis and gluconeogenesis. In addition, may also function as scaffolding protein. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14921 MDa |
-Macromolecule #1: Fructose-bisphosphate aldolase A
| Macromolecule | Name: Fructose-bisphosphate aldolase A / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: fructose-bisphosphate aldolase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.394875 KDa |
| Sequence | String: MPHSHPALTP EQKKELSDIA HRIVAPGKGI LAADESTGSI AKRLQSIGTE NTEENRRFYR QLLLTADDRV NPCIGGVILF HETLYQKAD DGRPFPQVIK SKGGVVGIKV DKGVVPLAGT NGETTTQGLD GLSERCAQYK KDGADFAKWR CVLKIGEHTP S ALAIMENA ...String: MPHSHPALTP EQKKELSDIA HRIVAPGKGI LAADESTGSI AKRLQSIGTE NTEENRRFYR QLLLTADDRV NPCIGGVILF HETLYQKAD DGRPFPQVIK SKGGVVGIKV DKGVVPLAGT NGETTTQGLD GLSERCAQYK KDGADFAKWR CVLKIGEHTP S ALAIMENA NVLARYASIC QQNGIVPIVE PEILPDGDHD LKRCQYVTEK VLAAVYKALS DHHIYLEGTL LKPNMVTPGH AC TQKYSHE EIAMATVTAL RRTVPPAVTG VTFLSGGQSE EEASINLNAI NKCPLLKPWA LTFSYGRALQ ASALKAWGGK KEN LKAAQE EYVKRALANS LACQGKYTPS GQAGAAASES LFISNHAY UniProtKB: Fructose-bisphosphate aldolase A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: DTT are added freshly before use. | ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: OTHER | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: OTHER / Digitization - Dimensions - Width: 8192 pixel / Digitization - Dimensions - Height: 8192 pixel / Number grids imaged: 1 / Number real images: 8682 / Average exposure time: 1.347 sec. / Average electron dose: 60.0 e/Å2 Details: Images were collected using Direct Electron Apollo camera at a fixed movie frame rate of 60 frames/second. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 80.0 µm / Calibrated magnification: 72621 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)