[English] 日本語
 Yorodumi
Yorodumi- EMDB-28034: Cryo-EM structure of the Hermes transposase bound to two left-end... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Hermes transposase bound to two left-ends of its DNA transposon | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transposase / transpososome / BED domain / protein-DNA complex / RECOMBINATION / RECOMBINATION-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein dimerization activity / regulation of transcription by RNA polymerase II / DNA binding / zinc ion binding / nucleus Similarity search - Function | |||||||||
| Biological species |  Musca domestica (house fly) Musca domestica (house fly) | |||||||||
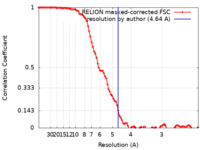
| Method | single particle reconstruction / cryo EM / Resolution: 4.64 Å | |||||||||
 Authors Authors | Lannes L / Dyda F | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Zinc-finger BED domains drive the formation of the active Hermes transpososome by asymmetric DNA binding. Authors: Laurie Lannes / Christopher M Furman / Alison B Hickman / Fred Dyda /  Abstract: The Hermes DNA transposon is a member of the eukaryotic hAT superfamily, and its transposase forms a ring-shaped tetramer of dimers. Our investigation, combining biochemical, crystallography and cryo- ...The Hermes DNA transposon is a member of the eukaryotic hAT superfamily, and its transposase forms a ring-shaped tetramer of dimers. Our investigation, combining biochemical, crystallography and cryo-electron microscopy, and in-cell assays, shows that the full-length Hermes octamer extensively interacts with its transposon left-end through multiple BED domains of three Hermes protomers contributed by three dimers explaining the role of the unusual higher-order assembly. By contrast, the right-end is bound to no BED domains at all. Thus, this work supports a model in which Hermes multimerizes to gather enough BED domains to find its left-end among the abundant genomic DNA, facilitating the subsequent interaction with the right-end. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_28034.map.gz emd_28034.map.gz | 65.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-28034-v30.xml emd-28034-v30.xml emd-28034.xml emd-28034.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_28034_fsc.xml emd_28034_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_28034.png emd_28034.png | 81.6 KB | ||
| Filedesc metadata |  emd-28034.cif.gz emd-28034.cif.gz | 6.9 KB | ||
| Others |  emd_28034_half_map_1.map.gz emd_28034_half_map_1.map.gz emd_28034_half_map_2.map.gz emd_28034_half_map_2.map.gz | 54.6 MB 54.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-28034 http://ftp.pdbj.org/pub/emdb/structures/EMD-28034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28034 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-28034 | HTTPS FTP |
-Related structure data
| Related structure data |  8edgMC  8eb5C  8sjdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_28034.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_28034.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.16 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_28034_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_28034_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Two left-end Hermes transpososome
| Entire | Name: Two left-end Hermes transpososome |
|---|---|
| Components |
|
-Supramolecule #1: Two left-end Hermes transpososome
| Supramolecule | Name: Two left-end Hermes transpososome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Hermes transposase tetramer of dimers complex bound to two transposon left-end DNAs. The complex was obtained by mixing the purified protein and the DNA. |
|---|---|
| Source (natural) | Organism:  Musca domestica (house fly) Musca domestica (house fly) |
| Molecular weight | Theoretical: 627 KDa |
-Macromolecule #1: DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3')
| Macromolecule | Name: DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3') / type: dna / ID: 1 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Musca domestica (house fly) Musca domestica (house fly) |
| Molecular weight | Theoretical: 2.162448 KDa |
| Sequence | String: (DG)(DC)(DG)(DT)(DG)(DA)(DA) |
-Macromolecule #2: DNA (46-MER)
| Macromolecule | Name: DNA (46-MER) / type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Musca domestica (house fly) Musca domestica (house fly) |
| Molecular weight | Theoretical: 14.173139 KDa |
| Sequence | String: (DA)(DG)(DA)(DG)(DA)(DA)(DC)(DA)(DA)(DC) (DA)(DA)(DC)(DA)(DA)(DG)(DT)(DG)(DG)(DC) (DT)(DT)(DA)(DT)(DT)(DT)(DT)(DG)(DA) (DT)(DA)(DC)(DT)(DT)(DA)(DT)(DG)(DC)(DG) (DC) (DC)(DA)(DC)(DT)(DT)(DG) |
-Macromolecule #3: DNA (55-MER)
| Macromolecule | Name: DNA (55-MER) / type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Musca domestica (house fly) Musca domestica (house fly) |
| Molecular weight | Theoretical: 16.931867 KDa |
| Sequence | String: (DC)(DA)(DA)(DG)(DT)(DG)(DG)(DC)(DG)(DC) (DA)(DT)(DA)(DA)(DG)(DT)(DA)(DT)(DC)(DA) (DA)(DA)(DA)(DT)(DA)(DA)(DG)(DC)(DC) (DA)(DC)(DT)(DT)(DG)(DT)(DT)(DG)(DT)(DT) (DG) (DT)(DT)(DC)(DT)(DC)(DT) ...String: (DC)(DA)(DA)(DG)(DT)(DG)(DG)(DC)(DG)(DC) (DA)(DT)(DA)(DA)(DG)(DT)(DA)(DT)(DC)(DA) (DA)(DA)(DA)(DT)(DA)(DA)(DG)(DC)(DC) (DA)(DC)(DT)(DT)(DG)(DT)(DT)(DG)(DT)(DT) (DG) (DT)(DT)(DC)(DT)(DC)(DT)(DG)(DG) (DT)(DT)(DC)(DA)(DC)(DG)(DC) |
-Macromolecule #4: Hermes transposase
| Macromolecule | Name: Hermes transposase / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Musca domestica (house fly) Musca domestica (house fly) |
| Molecular weight | Theoretical: 70.21057 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEKMDNLEVK AKINQGLYKI TPRHKGTSFI WNVLADIQKE DDTLVEGWVF CRKCEKVLKY TTRQTSNLCR HKCCASLKQS RELKTVSAD CKKEAIEKCA QWVVRDCRPF SAVSGSGFID MIKFFIKVGA EYGEHVNVEE LLPSPITLSR KVTSDAKEKK A LISREIKS ...String: MEKMDNLEVK AKINQGLYKI TPRHKGTSFI WNVLADIQKE DDTLVEGWVF CRKCEKVLKY TTRQTSNLCR HKCCASLKQS RELKTVSAD CKKEAIEKCA QWVVRDCRPF SAVSGSGFID MIKFFIKVGA EYGEHVNVEE LLPSPITLSR KVTSDAKEKK A LISREIKS AVEKDGASAT IDLWTDNYIK RNFLGVTLHY HENNELRDLI LGLKSLDFER STAENIYKKL KAIFSQFNVE DL SSIKFVT DRGANVVKSL ANNIRINCSS HLLSNVLENS FEETPELNMP ILACKNIVKY FKKANLQHRL RSSLKSECPT RWN STYTML RSILDNWESV IQILSEAGET QRIVHINKSI IQTMVNILDG FERIFKELQT CSSPSLCFVV PSILKVKEIC SPDV GDVAD IAKLKVNIIK NVRIIWEENL SIWHYTAFFF YPPALHMQQE KVAQIKEFCL SKMEDLELIN RMSSFNELSA TQLNQ SDSN SHNSIDLTSH SKDISTTSFF FPQLTQNNSR EPPVCPSDEF EFYRKEIVIL SEDFKVMEWW NLNSKKYPKL SKLALS LLS IPASSAASER TFSLAGNIIT EKRNRIGQQT VDSLLFLNSF YKNFCKLDI UniProtKB: Hermes transposase |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 6 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 2 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | The complex was formed in vitro by mixing the purified protein with the DNA and further purified by size exclusion chromatography. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 9500 / Average exposure time: 2.0 sec. / Average electron dose: 22.3 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||||
| Output model |  PDB-8edg: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)