[English] 日本語
 Yorodumi
Yorodumi- EMDB-27985: Yeast VO missing subunits a, e, and f in complex with Vma12-22p -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast VO missing subunits a, e, and f in complex with Vma12-22p | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | V-type proton ATPase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationVma12-Vma22 assembly complex / vacuolar proton-transporting V-type ATPase complex assembly / proton-transporting V-type ATPase, V1 domain / Insulin receptor recycling / Transferrin endocytosis and recycling / ROS and RNS production in phagocytes / Amino acids regulate mTORC1 / Golgi lumen acidification / P-type proton-exporting transporter activity / vacuolar transport ...Vma12-Vma22 assembly complex / vacuolar proton-transporting V-type ATPase complex assembly / proton-transporting V-type ATPase, V1 domain / Insulin receptor recycling / Transferrin endocytosis and recycling / ROS and RNS production in phagocytes / Amino acids regulate mTORC1 / Golgi lumen acidification / P-type proton-exporting transporter activity / vacuolar transport / vacuolar proton-transporting V-type ATPase, V0 domain / endosomal lumen acidification / vacuole organization / protein targeting to vacuole / proton-transporting V-type ATPase complex / fungal-type vacuole / vacuolar proton-transporting V-type ATPase complex / vacuolar acidification / fungal-type vacuole membrane / proton transmembrane transporter activity / proton-transporting ATPase activity, rotational mechanism / intracellular copper ion homeostasis / endomembrane system / Neutrophil degranulation / proton transmembrane transport / cell periphery / endocytosis / unfolded protein binding / protein-containing complex assembly / intracellular iron ion homeostasis / Golgi membrane / endoplasmic reticulum membrane / endoplasmic reticulum / nucleus / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Wang H / Bueler SA / Rubinstein JL | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Structural basis of V-ATPase V region assembly by Vma12p, 21p, and 22p. Authors: Hanlin Wang / Stephanie A Bueler / John L Rubinstein /  Abstract: Vacuolar-type adenosine triphosphatases (V-ATPases) are rotary proton pumps that acidify specific intracellular compartments in almost all eukaryotic cells. These multi-subunit enzymes consist of a ...Vacuolar-type adenosine triphosphatases (V-ATPases) are rotary proton pumps that acidify specific intracellular compartments in almost all eukaryotic cells. These multi-subunit enzymes consist of a soluble catalytic V region and a membrane-embedded proton-translocating V region. V is assembled in the endoplasmic reticulum (ER) membrane, and V is assembled in the cytosol. However, V binds V only after V is transported to the Golgi membrane, thereby preventing acidification of the ER. We isolated V complexes and subcomplexes from bound to V-ATPase assembly factors Vma12p, Vma21p, and Vma22p. Electron cryomicroscopy shows how the Vma12-22p complex recruits subunits a, e, and f to the rotor ring of V while blocking premature binding of V. Vma21p, which contains an ER-retrieval motif, binds the V:Vma12-22p complex, "mature" V, and a complex that appears to contain a ring of loosely packed rotor subunits and the proteins YAR027W and YAR028W. The structures suggest that Vma21p binds assembly intermediates that contain a rotor ring and that activation of proton pumping following assembly of V with V removes Vma21p, allowing V-ATPase to remain in the Golgi. Together, these structures show how Vma12-22p and Vma21p function in V-ATPase assembly and quality control, ensuring the enzyme acidifies only its intended cellular targets. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: Structural basis of V-ATPase V0 region assembly by Vma12p, 21p, and 22p Authors: Wang H / Bueler SA / Rubinstein JL | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_27985.map.gz emd_27985.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-27985-v30.xml emd-27985-v30.xml emd-27985.xml emd-27985.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_27985_fsc.xml emd_27985_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_27985.png emd_27985.png | 47.6 KB | ||
| Filedesc metadata |  emd-27985.cif.gz emd-27985.cif.gz | 6.8 KB | ||
| Others |  emd_27985_half_map_1.map.gz emd_27985_half_map_1.map.gz emd_27985_half_map_2.map.gz emd_27985_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-27985 http://ftp.pdbj.org/pub/emdb/structures/EMD-27985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27985 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-27985 | HTTPS FTP |
-Related structure data
| Related structure data |  8eatMC  8easC  8eauC  8eavC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_27985.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_27985.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_27985_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
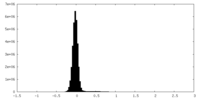
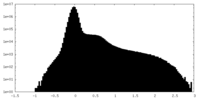
| Density Histograms |
-Half map: #2
| File | emd_27985_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
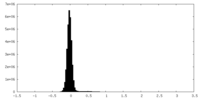
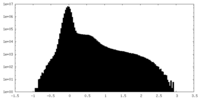
| Density Histograms |
- Sample components
Sample components
-Entire : Yeast VO missing subunits a, e, and f in complex with Vma12-22p
| Entire | Name: Yeast VO missing subunits a, e, and f in complex with Vma12-22p |
|---|---|
| Components |
|
-Supramolecule #1: Yeast VO missing subunits a, e, and f in complex with Vma12-22p
| Supramolecule | Name: Yeast VO missing subunits a, e, and f in complex with Vma12-22p type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Vacuolar ATPase assembly protein VMA22
| Macromolecule | Name: Vacuolar ATPase assembly protein VMA22 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.104717 KDa |
| Sequence | String: MSETRMAQNM DTTDEQYLRL IELLSNYDST LEQLQKGFQD GYIQLSRSNY YNKDSLRGNY GEDYWDETYI GQLMATVEEK NSKVVVEIV KRKAQDKQEK KEEEDNKLTQ RKKGTKPEKQ KTQSHKLKQD YDPILMFGGV LSVPSSLRQS QTSFKGCIPL I AQLINYKN EILTLVETLS EQE UniProtKB: Vacuolar ATPase assembly protein VMA22 |
-Macromolecule #2: Vacuolar ATPase assembly integral membrane protein VPH2
| Macromolecule | Name: Vacuolar ATPase assembly integral membrane protein VPH2 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.325648 KDa |
| Sequence | String: MFEIKLNDRI TEFLRKFKNS AKSNEGIDED IDLFLKRHAI PMQSLLFYVK EYRKDSDLQC SIKELLKPLE FEFKPKAVRG LHYSEDFKK KLEFLKYQEQ ELEYQSMVKR SKSVFSLQED DELTPSQINK QIKEQVTTVF NVLVSVISVV VAIWYWTGSS T NFPVHVRL ...String: MFEIKLNDRI TEFLRKFKNS AKSNEGIDED IDLFLKRHAI PMQSLLFYVK EYRKDSDLQC SIKELLKPLE FEFKPKAVRG LHYSEDFKK KLEFLKYQEQ ELEYQSMVKR SKSVFSLQED DELTPSQINK QIKEQVTTVF NVLVSVISVV VAIWYWTGSS T NFPVHVRL LLCLFFGILV LVADVVVYNS YLKKLEEAKV KEKTKVEKKK VLSKITL UniProtKB: Vacuolar ATPase assembly integral membrane protein VPH2 |
-Macromolecule #3: V-type proton ATPase subunit F
| Macromolecule | Name: V-type proton ATPase subunit F / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.47917 KDa |
| Sequence | String: MAEKRTLIAV IADEDTTTGL LLAGIGQITP ETQEKNFFVY QEGKTTKEEI TDKFNHFTEE RDDIAILLIN QHIAENIRAR VDSFTNAFP AILEIPSKDH PYDPEKDSVL KRVRKLFGE UniProtKB: V-type proton ATPase subunit F |
-Macromolecule #4: V0 assembly protein 1
| Macromolecule | Name: V0 assembly protein 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.694885 KDa |
| Sequence | String: MVFGQLYALF IFTLSCCISK TVQADSSKES SSFISFDKES NWDTISTISS TADVISSVDS AIAVFEFDNF SLLDNLMIDE EYPFFNRFF ANDVSLTVHD DSPLNISQSL SPIMEQFTVD ELPESASDLL YEYSLDDKSI VLFKFTSDAY DLKKLDEFID S CLSFLEDK ...String: MVFGQLYALF IFTLSCCISK TVQADSSKES SSFISFDKES NWDTISTISS TADVISSVDS AIAVFEFDNF SLLDNLMIDE EYPFFNRFF ANDVSLTVHD DSPLNISQSL SPIMEQFTVD ELPESASDLL YEYSLDDKSI VLFKFTSDAY DLKKLDEFID S CLSFLEDK SGDNLTVVIN SLGWAFEDED GDDEYATEET LSHHDNNKGK EGDDDILSSI WTEGLLMCLI VSALLLFILI VA LSWISNL DITYGALEKS TNPIKKNN UniProtKB: V0 assembly protein 1 |
-Macromolecule #5: V-type proton ATPase subunit c''
| Macromolecule | Name: V-type proton ATPase subunit c'' / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.610641 KDa |
| Sequence | String: MNKESKDDDM SLGKFSFSHF LYYLVLIVVI VYGLYKLFTG HGSDINFGKF LLRTSPYMWA NLGIALCVGL SVVGAAWGIF ITGSSMIGA GVRAPRITTK NLISIIFCEV VAIYGLIIAI VFSSKLTVAT AENMYSKSNL YTGYSLFWAG ITVGASNLIC G IAVGITGA ...String: MNKESKDDDM SLGKFSFSHF LYYLVLIVVI VYGLYKLFTG HGSDINFGKF LLRTSPYMWA NLGIALCVGL SVVGAAWGIF ITGSSMIGA GVRAPRITTK NLISIIFCEV VAIYGLIIAI VFSSKLTVAT AENMYSKSNL YTGYSLFWAG ITVGASNLIC G IAVGITGA TAAISDAADS ALFVKILVIE IFGSILGLLG LIVGLLMAGK ASEFQ UniProtKB: V-type proton ATPase subunit c'' |
-Macromolecule #6: V-type proton ATPase subunit d
| Macromolecule | Name: V-type proton ATPase subunit d / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.822484 KDa |
| Sequence | String: MEGVYFNIDN GFIEGVVRGY RNGLLSNNQY INLTQCDTLE DLKLQLSSTD YGNFLSSVSS ESLTTSLIQE YASSKLYHEF NYIRDQSSG STRKFMDYIT YGYMIDNVAL MITGTIHDRD KGEILQRCHP LGWFDTLPTL SVATDLESLY ETVLVDTPLA P YFKNCFDT ...String: MEGVYFNIDN GFIEGVVRGY RNGLLSNNQY INLTQCDTLE DLKLQLSSTD YGNFLSSVSS ESLTTSLIQE YASSKLYHEF NYIRDQSSG STRKFMDYIT YGYMIDNVAL MITGTIHDRD KGEILQRCHP LGWFDTLPTL SVATDLESLY ETVLVDTPLA P YFKNCFDT AEELDDMNIE IIRNKLYKAY LEDFYNFVTE EIPEPAKECM QTLLGFEADR RSINIALNSL QSSDIDPDLK SD LLPNIGK LYPLATFHLA QAQDFEGVRA ALANVYEYRG FLETGNLEDH FYQLEMELCR DAFTQQFAIS TVWAWMKSKE QEV RNITWI AECIAQNQRE RINNYISVY UniProtKB: V-type proton ATPase subunit d |
-Macromolecule #7: V-type proton ATPase subunit c
| Macromolecule | Name: V-type proton ATPase subunit c / type: protein_or_peptide / ID: 7 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.357501 KDa |
| Sequence | String: MTELCPVYAP FFGAIGCASA IIFTSLGAAY GTAKSGVGIC ATCVLRPDLL FKNIVPVIMA GIIAIYGLVV SVLVCYSLGQ KQALYTGFI QLGAGLSVGL SGLAAGFAIG IVGDAGVRGS SQQPRLFVGM ILILIFAEVL GLYGLIVALL LNSRATQDVV C UniProtKB: V-type proton ATPase subunit c |
-Macromolecule #8: V-type proton ATPase subunit c'
| Macromolecule | Name: V-type proton ATPase subunit c' / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.046361 KDa |
| Sequence | String: MSTQLASNIY APLYAPFFGF AGCAAAMVLS CLGAAIGTAK SGIGIAGIGT FKPELIMKSL IPVVMSGILA IYGLVVAVLI AGNLSPTED YTLFNGFMHL SCGLCVGFAC LSSGYAIGMV GDVGVRKYMH QPRLFVGIVL ILIFSEVLGL YGMIVALILN T RGSE UniProtKB: V-type proton ATPase subunit c' |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Homemade / Material: COPPER/RHODIUM |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)