[English] 日本語
 Yorodumi
Yorodumi- EMDB-26665: Tau Paired Helical Filament from Alzheimer's Disease incubated wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tau Paired Helical Filament from Alzheimer's Disease incubated with EGCG for 3 hours | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Amyloid / fibril / Alzheimer's / disease / disaggregant / PROTEIN FIBRIL | |||||||||||||||
| Function / homology | Activation of AMPK downstream of NMDARs / PKR-mediated signaling / Isoform Tau-F of Microtubule-associated protein tau Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||||||||
 Authors Authors | Seidler PM / Murray KA / Boyer DR / Ge P / Sawaya MR / Eisenberg DS | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structure-based discovery of small molecules that disaggregate Alzheimer's disease tissue derived tau fibrils in vitro. Authors: Paul M Seidler / Kevin A Murray / David R Boyer / Peng Ge / Michael R Sawaya / Carolyn J Hu / Xinyi Cheng / Romany Abskharon / Hope Pan / Michael A DeTure / Christopher K Williams / Dennis W ...Authors: Paul M Seidler / Kevin A Murray / David R Boyer / Peng Ge / Michael R Sawaya / Carolyn J Hu / Xinyi Cheng / Romany Abskharon / Hope Pan / Michael A DeTure / Christopher K Williams / Dennis W Dickson / Harry V Vinters / David S Eisenberg /  Abstract: Alzheimer's disease (AD) is the consequence of neuronal death and brain atrophy associated with the aggregation of protein tau into fibrils. Thus disaggregation of tau fibrils could be a therapeutic ...Alzheimer's disease (AD) is the consequence of neuronal death and brain atrophy associated with the aggregation of protein tau into fibrils. Thus disaggregation of tau fibrils could be a therapeutic approach to AD. The small molecule EGCG, abundant in green tea, has long been known to disaggregate tau and other amyloid fibrils, but EGCG has poor drug-like properties, failing to fully penetrate the brain. Here we have cryogenically trapped an intermediate of brain-extracted tau fibrils on the kinetic pathway to EGCG-induced disaggregation and have determined its cryoEM structure. The structure reveals that EGCG molecules stack in polar clefts between the paired helical protofilaments that pathologically define AD. Treating the EGCG binding position as a pharmacophore, we computationally screened thousands of drug-like compounds for compatibility for the pharmacophore, discovering several that experimentally disaggregate brain-derived tau fibrils in vitro. This work suggests the potential of structure-based, small-molecule drug discovery for amyloid diseases. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26665.map.gz emd_26665.map.gz | 35.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26665-v30.xml emd-26665-v30.xml emd-26665.xml emd-26665.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_26665.png emd_26665.png | 82.7 KB | ||
| Filedesc metadata |  emd-26665.cif.gz emd-26665.cif.gz | 5.7 KB | ||
| Others |  emd_26665_half_map_1.map.gz emd_26665_half_map_1.map.gz emd_26665_half_map_2.map.gz emd_26665_half_map_2.map.gz | 285.7 MB 285.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26665 http://ftp.pdbj.org/pub/emdb/structures/EMD-26665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26665 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26665 | HTTPS FTP |
-Validation report
| Summary document |  emd_26665_validation.pdf.gz emd_26665_validation.pdf.gz | 976.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_26665_full_validation.pdf.gz emd_26665_full_validation.pdf.gz | 976 KB | Display | |
| Data in XML |  emd_26665_validation.xml.gz emd_26665_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_26665_validation.cif.gz emd_26665_validation.cif.gz | 18.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26665 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26665 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26665 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-26665 | HTTPS FTP |
-Related structure data
| Related structure data |  7upgMC  7upeC  7upfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26665.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26665.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.064 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_26665_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_26665_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tau Paired Helical Filament from Alzheimer's Disease incubated wi...
| Entire | Name: Tau Paired Helical Filament from Alzheimer's Disease incubated with EGCG for 3 hours |
|---|---|
| Components |
|
-Supramolecule #1: Tau Paired Helical Filament from Alzheimer's Disease incubated wi...
| Supramolecule | Name: Tau Paired Helical Filament from Alzheimer's Disease incubated with EGCG for 3 hours type: tissue / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform Tau-F of Microtubule-associated protein tau
| Macromolecule | Name: Isoform Tau-F of Microtubule-associated protein tau / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.919871 KDa |
| Sequence | String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKESPLQT PTEDGSEEPG SETSDAKSTP TAEDVTAPLV DEGAPGKQA AAQPHTEIPE GTTAEEAGIG DTPSLEDEAA GHVTQARMVS KSKDGTGSDD KKAKGADGKT KIATPRGAAP P GQKGQANA ...String: MAEPRQEFEV MEDHAGTYGL GDRKDQGGYT MHQDQEGDTD AGLKESPLQT PTEDGSEEPG SETSDAKSTP TAEDVTAPLV DEGAPGKQA AAQPHTEIPE GTTAEEAGIG DTPSLEDEAA GHVTQARMVS KSKDGTGSDD KKAKGADGKT KIATPRGAAP P GQKGQANA TRIPAKTPPA PKTPPSSGEP PKSGDRSGYS SPGSPGTPGS RSRTPSLPTP PTREPKKVAV VRTPPKSPSS AK SRLQTAP VPMPDLKNVK SKIGSTENLK HQPGGGKVQI INKKLDLSNV QSKCGSKDNI KHVPGGGSVQ IVYKPVDLSK VTS KCGSLG NIHHKPGGGQ VEVKSEKLDF KDRVQSKIGS LDNITHVPGG GNKKIETHKL TFRENAKAKT DHGAEIVYKS PVVS GDTSP RHLSNVSSTG SIDMVDSPQL ATLADEVSAS LAKQGL UniProtKB: Isoform Tau-F of Microtubule-associated protein tau |
-Macromolecule #2: (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-c...
| Macromolecule | Name: (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate type: ligand / ID: 2 / Number of copies: 10 / Formula: KDH |
|---|---|
| Molecular weight | Theoretical: 458.372 Da |
| Chemical component information | 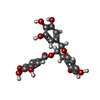 ChemComp-EGG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 6988 / Average electron dose: 0.66 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 2.4 Å Applied symmetry - Helical parameters - Δ&Phi: 179.46 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 29145 |
|---|---|
| Startup model | Type of model: OTHER Details: Featureless gaussian cylinder generated from relion_helix_toolbox. |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





































