[English] 日本語
 Yorodumi
Yorodumi- EMDB-25165: Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the PR-RT components of the HIV-1 Pol Polyprotein | ||||||||||||
 Map data Map data | Full-map generated by Focussed Classification filtered by 2 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | HIV-1 / Reverse transcriptase / Protease / VIRAL PROTEIN / enzyme | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationHIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding ...HIV-1 retropepsin / symbiont-mediated activation of host apoptosis / retroviral ribonuclease H / exoribonuclease H / exoribonuclease H activity / DNA integration / viral genome integration into host DNA / establishment of integrated proviral latency / RNA-directed DNA polymerase / RNA stem-loop binding / viral penetration into host nucleus / host multivesicular body / RNA-directed DNA polymerase activity / RNA-DNA hybrid ribonuclease activity / Transferases; Transferring phosphorus-containing groups; Nucleotidyltransferases / host cell / viral nucleocapsid / DNA recombination / DNA-directed DNA polymerase / aspartic-type endopeptidase activity / Hydrolases; Acting on ester bonds / DNA-directed DNA polymerase activity / symbiont-mediated suppression of host gene expression / viral translational frameshifting / symbiont entry into host cell / lipid binding / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / DNA binding / zinc ion binding Similarity search - Function | ||||||||||||
| Biological species |  Human immunodeficiency virus type 1 BH10 / Human immunodeficiency virus type 1 BH10 /  Human immunodeficiency virus type 1 group M subtype B (isolate BH10) Human immunodeficiency virus type 1 group M subtype B (isolate BH10) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.2 Å | ||||||||||||
 Authors Authors | Lyumkis D / Passos D | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation. Authors: Jerry Joe E K Harrison / Dario Oliveira Passos / Jessica F Bruhn / Joseph D Bauman / Lynda Tuberty / Jeffrey J DeStefano / Francesc Xavier Ruiz / Dmitry Lyumkis / Eddy Arnold /  Abstract: Key proteins of retroviruses and other RNA viruses are translated and subsequently processed from polyprotein precursors by the viral protease (PR). Processing of the HIV Gag-Pol polyprotein yields ...Key proteins of retroviruses and other RNA viruses are translated and subsequently processed from polyprotein precursors by the viral protease (PR). Processing of the HIV Gag-Pol polyprotein yields the HIV structural proteins and enzymes. Structures of the mature enzymes PR, reverse transcriptase (RT), and integrase (IN) aided understanding of catalysis and design of antiretrovirals, but knowledge of the Pol precursor architecture and function before PR cleavage is limited. We developed a system to produce stable HIV-1 Pol and determined its cryo-electron microscopy structure. RT in Pol has a similar arrangement to the mature RT heterodimer, and its dimerization may draw together two PR monomers to activate proteolytic processing. HIV-1 thus may leverage the dimerization interfaces in Pol to regulate assembly and maturation of polyprotein precursors. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25165.map.gz emd_25165.map.gz | 59.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25165-v30.xml emd-25165-v30.xml emd-25165.xml emd-25165.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_25165.png emd_25165.png | 114 KB | ||
| Others |  emd_25165_additional_1.map.gz emd_25165_additional_1.map.gz emd_25165_half_map_1.map.gz emd_25165_half_map_1.map.gz emd_25165_half_map_2.map.gz emd_25165_half_map_2.map.gz | 59.3 MB 12.4 MB 12.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25165 http://ftp.pdbj.org/pub/emdb/structures/EMD-25165 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25165 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25165 | HTTPS FTP |
-Related structure data
| Related structure data |  7sjxMC  7sepC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25165.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25165.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full-map generated by Focussed Classification filtered by 2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.015 Å | ||||||||||||||||||||||||||||||||||||
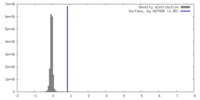
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Full-map generated by Focussed Classification
| File | emd_25165_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full-map generated by Focussed Classification | ||||||||||||
| Projections & Slices |
| ||||||||||||
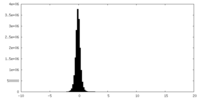
| Density Histograms |
-Half map: Half-map 2
| File | emd_25165_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
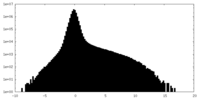
| Density Histograms |
-Half map: Half-map 1
| File | emd_25165_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
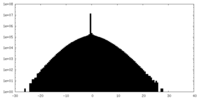
| Density Histograms |
- Sample components
Sample components
-Entire : PR-RT portion of HIV-1 Pol
| Entire | Name: PR-RT portion of HIV-1 Pol |
|---|---|
| Components |
|
-Supramolecule #1: PR-RT portion of HIV-1 Pol
| Supramolecule | Name: PR-RT portion of HIV-1 Pol / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: 3D reconstruction of the HIV-1 Pol polyprotein comprising the PR-RT portion |
|---|---|
| Source (natural) | Organism:  Human immunodeficiency virus type 1 BH10 Human immunodeficiency virus type 1 BH10 |
| Molecular weight | Theoretical: 130 KDa |
-Macromolecule #1: Gag-Pol polyprotein
| Macromolecule | Name: Gag-Pol polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human immunodeficiency virus type 1 group M subtype B (isolate BH10) Human immunodeficiency virus type 1 group M subtype B (isolate BH10)Strain: isolate BH10 |
| Molecular weight | Theoretical: 119.289867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH MATVKFKYKG EEKEVDISKI KKVWRVGKMI SFTYDEGGGK TGRGAVSEKD APKELLQMLE KQKKALEVLF QGPMGRDNN SPSEAGADRQ GTVSFNFPQI TLWQRPLVTI KIGGQLKEAL LATGADDTVL EEMSLPGRWK PKMIGGIGGF I KVRQYDQI ...String: MGSSHHHHHH MATVKFKYKG EEKEVDISKI KKVWRVGKMI SFTYDEGGGK TGRGAVSEKD APKELLQMLE KQKKALEVLF QGPMGRDNN SPSEAGADRQ GTVSFNFPQI TLWQRPLVTI KIGGQLKEAL LATGADDTVL EEMSLPGRWK PKMIGGIGGF I KVRQYDQI LIEICGHKAI GTVLVGPTPV NIIGRNLLTQ IGCTLNFPIS PIETVPVKLK PGMDGPKVKQ WPLTEEKIKA LV EICTEME KEGKISKIGP ENPYNTPVFA IKKKDSTKWR KLVDFRELNK RTQDFWEVQL GIPHPAGLKK KKSVTVLDVG DAY FSVPLD EDFRKYTAFT IPSINNETPG IRYQYNVLPQ GWKGSPAIFQ SSMTKILEPF KKQNPDIVIY QYMDDLYVGS DLEI GQHRT KIEELRQHLL RWGLTTPDKK HQKEPPFLWM GYELHPDKWT VQPIVLPEKD SWTVNDIQKL VGKLNWASQI YPGIK VRQL CKLLRGTKAL TEVIPLTEEA ELELAENREI LKEPVHGVYY DPSKDLIAEI QKQGQGQWTY QIYQEPFKNL KTGKYA RMR GAHTNDVKQL TEAVQKITTE SIVIWGKTPK FKLPIQKETW ETWWTEYWQA TWIPEWEFVN TPPLVKLWYQ LEKEPIV GA ETFYVDGAAN RETKLGKAGY VTNKGRQKVV PLTNTTNQKT ELQAIYLALQ DSGLEVNIVT DSQYALGIIQ AQPDKSES E LVNQIIEQLI KKEKVYLAWV PAHKGIGGNE QVDKLVSAGI RKIDDLDGID KAQDEHEKYH SNWRAMASDF NLPPVVAKE IVASCDKCQL KGEAMHGQVD CSPGIWQLDC THLEGKVILV AVHVASGYIE AEVIPAETGQ ETAYFLLKLA GRWPVKTIHT DNGSNFTSA TVKAACWWAG IKQEFGIPYN PQSQGVVESM NKELKKIIGQ VRDQAEHLKT AVQMAVFIHN FKRKGGIGGY S AGERIVDI IATDIQTKEL QKQITKIQNF RVYYRDSRNP LWKGPAKLLW KGEGAVVIQD NSDIKVVPRR KAKIIRDYGK QM AGDDCVA SRQDED UniProtKB: Gag-Pol polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Average electron dose: 0.95 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)