+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24444 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse GITR (mGITR) with DTA-1 Fab fragment | |||||||||
 Map data Map data | mGITR with DTA-1 Fab | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mouse glucocorticoid-induced tumor necrosis factor receptor / mGITR / TNF receptor / DTA-1 / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationTNFs bind their physiological receptors / tumor necrosis factor receptor activity / positive regulation of cell adhesion / bioluminescence / generation of precursor metabolites and energy / external side of plasma membrane / apoptotic process / negative regulation of apoptotic process / extracellular region Similarity search - Function | |||||||||
| Biological species |   Muromegalovirus G4 Muromegalovirus G4 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Meyerson JR / He C | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Therapeutic antibody activation of the glucocorticoid-induced TNF receptor by a clustering mechanism. Authors: Changhao He / Rachana R Maniyar / Yahel Avraham / Roberta Zappasodi / Radda Rusinova / Walter Newman / Heidi Heath / Jedd D Wolchok / Rony Dahan / Taha Merghoub / Joel R Meyerson /   Abstract: GITR is a TNF receptor, and its activation promotes immune responses and drives antitumor activity. The receptor is activated by the GITR ligand (GITRL), which is believed to cluster receptors into a ...GITR is a TNF receptor, and its activation promotes immune responses and drives antitumor activity. The receptor is activated by the GITR ligand (GITRL), which is believed to cluster receptors into a high-order array. Immunotherapeutic agonist antibodies also activate the receptor, but their mechanisms are not well characterized. We solved the structure of full-length mouse GITR bound to Fabs from the antibody DTA-1. The receptor is a dimer, and each subunit binds one Fab in an orientation suggesting that the antibody clusters receptors. Binding experiments with purified proteins show that DTA-1 IgG and GITRL both drive extensive clustering of GITR. Functional data reveal that DTA-1 and the anti-human GITR antibody TRX518 activate GITR in their IgG forms but not as Fabs. Thus, the divalent character of the IgG agonists confers an ability to mimic GITRL and cluster and activate GITR. These findings will inform the clinical development of this class of antibodies for immuno-oncology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24444.map.gz emd_24444.map.gz | 203.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24444-v30.xml emd-24444-v30.xml emd-24444.xml emd-24444.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
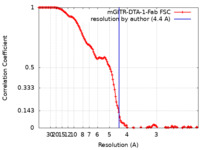
| FSC (resolution estimation) |  emd_24444_fsc.xml emd_24444_fsc.xml | 13.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_24444.png emd_24444.png | 62.2 KB | ||
| Filedesc metadata |  emd-24444.cif.gz emd-24444.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24444 http://ftp.pdbj.org/pub/emdb/structures/EMD-24444 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24444 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24444 | HTTPS FTP |
-Related structure data
| Related structure data |  7rfpMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24444.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24444.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mGITR with DTA-1 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.096 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : mGITR with DTA-1 Fab
| Entire | Name: mGITR with DTA-1 Fab |
|---|---|
| Components |
|
-Supramolecule #1: mGITR with DTA-1 Fab
| Supramolecule | Name: mGITR with DTA-1 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tumor necrosis factor receptor superfamily member 18,Enhanced gre...
| Macromolecule | Name: Tumor necrosis factor receptor superfamily member 18,Enhanced green fluorescent protein type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Muromegalovirus G4 Muromegalovirus G4 |
| Molecular weight | Theoretical: 54.848238 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGAWAMLYGV SMLCVLDLGQ PSVVEEPGCG PGKVQNGSGN NTRCCSLYAP GKEDCPKERC ICVTPEYHCG DPQCKICKHY PCQPGQRVE SQGDIVFGFR CVACAMGTFS AGRDGHCRLW TNCSQFGFLT MFPGNKTHNA VCIPEPLPTE QYGHLTVIFL V MAACIFFL ...String: MGAWAMLYGV SMLCVLDLGQ PSVVEEPGCG PGKVQNGSGN NTRCCSLYAP GKEDCPKERC ICVTPEYHCG DPQCKICKHY PCQPGQRVE SQGDIVFGFR CVACAMGTFS AGRDGHCRLW TNCSQFGFLT MFPGNKTHNA VCIPEPLPTE QYGHLTVIFL V MAACIFFL TTVQLGLHIW QLRRQHMCPR ETQPFAEVQL SAEDACSFQF PEEERGEQTE EKCHLGGRWP AGLVPRGSAA AM VSKGEEL FTGVVPILVE LDGDVNGHKF SVSGEGEGDA TYGKLTLKFI CTTGKLPVPW PTLVTTLTYG VQCFSRYPDH MKQ HDFFKS AMPEGYVQER TIFFKDDGNY KTRAEVKFEG DTLVNRIELK GIDFKEDGNI LGHKLEYNYN SHNVYIMADK QKNG IKVNF KIRHNIEDGS VQLADHYQQN TPIGDGPVLL PDNHYLSTQS KLSKDPNEKR DHMVLLEFVT AAGITLGMDE LYKSG LRHH HHHHHH UniProtKB: Tumor necrosis factor receptor superfamily member 18, Enhanced green fluorescent protein |
-Macromolecule #2: DTA-1 (heavy chain)
| Macromolecule | Name: DTA-1 (heavy chain) / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.014621 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSCIILFL VATATGVHSE LQLVESGGGL VQPGRSLKLS CSASGFIFSN SYMAWVRQAP KKGLEWVATI NPSGSRTYYP DSVKGRFTI SRDTAKSSLY LQMNSLKSED TATYYCARHD LSFDYWGQGV MVTVSSASTK GPSVFPLAPS SKSTSGGTAA L GCLVKDYF ...String: MGWSCIILFL VATATGVHSE LQLVESGGGL VQPGRSLKLS CSASGFIFSN SYMAWVRQAP KKGLEWVATI NPSGSRTYYP DSVKGRFTI SRDTAKSSLY LQMNSLKSED TATYYCARHD LSFDYWGQGV MVTVSSASTK GPSVFPLAPS SKSTSGGTAA L GCLVKDYF PEPVTVSWNS GALTSGVHTF PAVLQSSGLY SLSSVVTVPS SSLGTQTYIC NVNHKPSNTK VDKRVEPKSC DK THTCPPC PAPELLGGPS VFLFPPKPKD TLMISRTPEV TCVVVDVSHE DPEVKFNWYV DGVEVHNAKT KPREEQYNST YRV VSVLTV LHQDWLNGKE YKCKVSNKAL PAPIEKTISK AKGQPREPQV YTLPPSREEM TKNQVSLTCL VKGFYPSDIA VEWE SNGQP ENNYKTTPPV LDSDGSFFLY SKLTVDKSRW QQGNVFSCSV MHEALHNHYT QKSLSLSPGK |
-Macromolecule #3: DTA-1 (light chain)
| Macromolecule | Name: DTA-1 (light chain) / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.320057 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSCIILFL VATATGVHSQ FTLTQPKSVS GSLRSTITIP CDRSSGGIRD SYVSWYQQHL GRPPLNVIYA DDQRPSEVSD RFSGSIDSS SNSASLTITN LQMDDEADYF CQSYDSDFDV YIFGGGTKLT VLGQRTVAAP SVFIFPPSDE QLKSGTASVV C LLNNFYPR ...String: MGWSCIILFL VATATGVHSQ FTLTQPKSVS GSLRSTITIP CDRSSGGIRD SYVSWYQQHL GRPPLNVIYA DDQRPSEVSD RFSGSIDSS SNSASLTITN LQMDDEADYF CQSYDSDFDV YIFGGGTKLT VLGQRTVAAP SVFIFPPSDE QLKSGTASVV C LLNNFYPR EAKVQWKVDN ALQSGNSQES VTEQDSKDST YSLSSTLTLS KADYEKHKVY ACEVTHQGLS SPVTKSFNRG EC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)