[English] 日本語
 Yorodumi
Yorodumi- EMDB-24272: Cryo-EM structure of activated human SARM1 in complex with NMN an... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of activated human SARM1 in complex with NMN and 1AD (TIR:1AD) | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NADase / axon degeneration / inhibitor / HYDROLASE / HYDROLASE-Inhibitor complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationextrinsic component of synaptic membrane / negative regulation of MyD88-independent toll-like receptor signaling pathway / NADP+ nucleosidase activity / MyD88-independent TLR4 cascade / Toll Like Receptor 3 (TLR3) Cascade / NAD+ catabolic process / NAD+ nucleosidase activity / regulation of synapse pruning / modification of postsynaptic structure / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase ...extrinsic component of synaptic membrane / negative regulation of MyD88-independent toll-like receptor signaling pathway / NADP+ nucleosidase activity / MyD88-independent TLR4 cascade / Toll Like Receptor 3 (TLR3) Cascade / NAD+ catabolic process / NAD+ nucleosidase activity / regulation of synapse pruning / modification of postsynaptic structure / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / protein localization to mitochondrion / NAD+ nucleosidase activity, cyclic ADP-ribose generating / nervous system process / Hydrolases; Glycosylases; Hydrolysing N-glycosyl compounds / regulation of dendrite morphogenesis / response to glucose / response to axon injury / regulation of neuron apoptotic process / signaling adaptor activity / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / IKK complex recruitment mediated by RIP1 / neuromuscular junction / nervous system development / microtubule / mitochondrial outer membrane / cell differentiation / axon / innate immune response / synapse / dendrite / glutamatergic synapse / cell surface / signal transduction / protein-containing complex / mitochondrion / identical protein binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
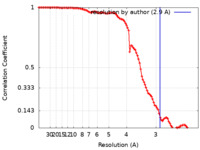
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Kerry PS / Nanson JD | |||||||||
| Funding support |  Australia, 2 items Australia, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules. Authors: Yun Shi / Philip S Kerry / Jeffrey D Nanson / Todd Bosanac / Yo Sasaki / Raul Krauss / Forhad K Saikot / Sarah E Adams / Tamim Mosaiab / Veronika Masic / Xianrong Mao / Faith Rose / Eduardo ...Authors: Yun Shi / Philip S Kerry / Jeffrey D Nanson / Todd Bosanac / Yo Sasaki / Raul Krauss / Forhad K Saikot / Sarah E Adams / Tamim Mosaiab / Veronika Masic / Xianrong Mao / Faith Rose / Eduardo Vasquez / Marieke Furrer / Katie Cunnea / Andrew Brearley / Weixi Gu / Zhenyao Luo / Lou Brillault / Michael J Landsberg / Aaron DiAntonio / Bostjan Kobe / Jeffrey Milbrandt / Robert O Hughes / Thomas Ve /     Abstract: The NADase SARM1 (sterile alpha and TIR motif containing 1) is a key executioner of axon degeneration and a therapeutic target for several neurodegenerative conditions. We show that a potent SARM1 ...The NADase SARM1 (sterile alpha and TIR motif containing 1) is a key executioner of axon degeneration and a therapeutic target for several neurodegenerative conditions. We show that a potent SARM1 inhibitor undergoes base exchange with the nicotinamide moiety of nicotinamide adenine dinucleotide (NAD) to produce the bona fide inhibitor 1AD. We report structures of SARM1 in complex with 1AD, NAD mimetics and the allosteric activator nicotinamide mononucleotide (NMN). NMN binding triggers reorientation of the armadillo repeat (ARM) domains, which disrupts ARM:TIR interactions and leads to formation of a two-stranded TIR domain assembly. The active site spans two molecules in these assemblies, explaining the requirement of TIR domain self-association for NADase activity and axon degeneration. Our results reveal the mechanisms of SARM1 activation and substrate binding, providing rational avenues for the design of new therapeutics targeting SARM1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24272.map.gz emd_24272.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24272-v30.xml emd-24272-v30.xml emd-24272.xml emd-24272.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24272_fsc.xml emd_24272_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_24272.png emd_24272.png | 98.5 KB | ||
| Masks |  emd_24272_msk_1.map emd_24272_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-24272.cif.gz emd-24272.cif.gz | 6.7 KB | ||
| Others |  emd_24272_half_map_1.map.gz emd_24272_half_map_1.map.gz emd_24272_half_map_2.map.gz emd_24272_half_map_2.map.gz | 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24272 http://ftp.pdbj.org/pub/emdb/structures/EMD-24272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24272 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24272 | HTTPS FTP |
-Validation report
| Summary document |  emd_24272_validation.pdf.gz emd_24272_validation.pdf.gz | 695.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24272_full_validation.pdf.gz emd_24272_full_validation.pdf.gz | 695.1 KB | Display | |
| Data in XML |  emd_24272_validation.xml.gz emd_24272_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  emd_24272_validation.cif.gz emd_24272_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24272 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24272 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24272 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24272 | HTTPS FTP |
-Related structure data
| Related structure data |  7nakMC  7nagC  7nahC  7naiC 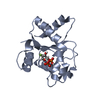 7najC  7nalC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24272.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24272.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.086 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_24272_msk_1.map emd_24272_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_24272_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B
| File | emd_24272_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : human SARM1 in complex with NMN and 1AD
| Entire | Name: human SARM1 in complex with NMN and 1AD |
|---|---|
| Components |
|
-Supramolecule #1: human SARM1 in complex with NMN and 1AD
| Supramolecule | Name: human SARM1 in complex with NMN and 1AD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: NAD(+) hydrolase SARM1
| Macromolecule | Name: NAD(+) hydrolase SARM1 / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO / EC number: ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 76.471469 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LAVPGPDGGG GTGPWWAAGG RGPREVSPGA GTEVQDALER ALPELQQALS ALKQAGGARA VGAGLAEVFQ LVEEAWLLPA VGREVAQGL CDAIRLDGGL DLLLRLLQAP ELETRVQAAR LLEQILVAEN RDRVARIGLG VILNLAKERE PVELARSVAG I LEHMFKHS ...String: LAVPGPDGGG GTGPWWAAGG RGPREVSPGA GTEVQDALER ALPELQQALS ALKQAGGARA VGAGLAEVFQ LVEEAWLLPA VGREVAQGL CDAIRLDGGL DLLLRLLQAP ELETRVQAAR LLEQILVAEN RDRVARIGLG VILNLAKERE PVELARSVAG I LEHMFKHS EETCQRLVAA GGLDAVLYWC RRTDPALLRH CALALGNCAL HGGQAVQRRM VEKRAAEWLF PLAFSKEDEL LR LHACLAV AVLATNKEVE REVERSGTLA LVEPLVASLD PGRFARCLVD ASDTSQGRGP DDLQRLVPLL DSNRLEAQCI GAF YLCAEA AIKSLQGKTK VFSDIGAIQS LKRLVSYSTN GTKSALAKRA LRLLGEEVPR PILPSVPSWK EAEVQTWLQQ IGFS KYCES FREQQVDGDL LLRLTEEELQ TDLGMKSGIT RKRFFRELTE LKTFANYSTC DRSNLADWLG SLDPRFRQYT YGLVS CGLD RSLLHRVSEQ QLLEDCGIHL GVHRARILTA AREMLHSPLP CTGGKPSGDT PDVFISYRRN SGSQLASLLK VHLQLH GFS VFIDVEKLEA GKFEDKLIQS VMGARNFVLV LSPGALDKCM QDHDCKDWVH KEIVTALSCG KNIVPIIDGF EWPEPQV LP EDMQAVLTFN GIKWSHEYQE ATIEKIIRFL QGRSSRDSSA GSDTSLEGAA PMGPT UniProtKB: NAD(+) hydrolase SARM1 |
-Macromolecule #2: [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidany...
| Macromolecule | Name: [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl] ...Name: [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(5-iodanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate type: ligand / ID: 2 / Number of copies: 8 / Formula: 1QD |
|---|---|
| Molecular weight | Theoretical: 797.364 Da |
| Chemical component information | 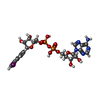 ChemComp-1QD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)