[English] 日本語
 Yorodumi
Yorodumi- PDB-7nah: Crystal structure of the TIR domain from human SARM1 in complex w... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7nah | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
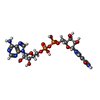
| Title | Crystal structure of the TIR domain from human SARM1 in complex with 2AD | |||||||||
 Components Components | Sterile alpha and TIR motif-containing protein 1 | |||||||||
 Keywords Keywords | HYDROLASE / NADase / Axon degeneration | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of MyD88-independent toll-like receptor signaling pathway / extrinsic component of synaptic membrane / MyD88-independent TLR4 cascade / NADP+ nucleosidase activity / Toll Like Receptor 3 (TLR3) Cascade / NAD+ catabolic process / NAD+ nucleosidase activity / regulation of synapse pruning / modification of postsynaptic structure / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase ...negative regulation of MyD88-independent toll-like receptor signaling pathway / extrinsic component of synaptic membrane / MyD88-independent TLR4 cascade / NADP+ nucleosidase activity / Toll Like Receptor 3 (TLR3) Cascade / NAD+ catabolic process / NAD+ nucleosidase activity / regulation of synapse pruning / modification of postsynaptic structure / ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase / protein localization to mitochondrion / NAD+ nucleosidase activity, cyclic ADP-ribose generating / nervous system process / Hydrolases; Glycosylases; Hydrolysing N-glycosyl compounds / regulation of dendrite morphogenesis / response to glucose / response to axon injury / signaling adaptor activity / TRAF6-mediated induction of TAK1 complex within TLR4 complex / regulation of neuron apoptotic process / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / IKK complex recruitment mediated by RIP1 / neuromuscular junction / nervous system development / microtubule / mitochondrial outer membrane / cell differentiation / axon / innate immune response / dendrite / synapse / glutamatergic synapse / cell surface / signal transduction / protein-containing complex / mitochondrion / identical protein binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.79 Å MOLECULAR REPLACEMENT / Resolution: 1.79 Å | |||||||||
 Authors Authors | Shi, Y. / Ve, T. | |||||||||
| Funding support |  Australia, 2items Australia, 2items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules. Authors: Yun Shi / Philip S Kerry / Jeffrey D Nanson / Todd Bosanac / Yo Sasaki / Raul Krauss / Forhad K Saikot / Sarah E Adams / Tamim Mosaiab / Veronika Masic / Xianrong Mao / Faith Rose / Eduardo ...Authors: Yun Shi / Philip S Kerry / Jeffrey D Nanson / Todd Bosanac / Yo Sasaki / Raul Krauss / Forhad K Saikot / Sarah E Adams / Tamim Mosaiab / Veronika Masic / Xianrong Mao / Faith Rose / Eduardo Vasquez / Marieke Furrer / Katie Cunnea / Andrew Brearley / Weixi Gu / Zhenyao Luo / Lou Brillault / Michael J Landsberg / Aaron DiAntonio / Bostjan Kobe / Jeffrey Milbrandt / Robert O Hughes / Thomas Ve /     Abstract: The NADase SARM1 (sterile alpha and TIR motif containing 1) is a key executioner of axon degeneration and a therapeutic target for several neurodegenerative conditions. We show that a potent SARM1 ...The NADase SARM1 (sterile alpha and TIR motif containing 1) is a key executioner of axon degeneration and a therapeutic target for several neurodegenerative conditions. We show that a potent SARM1 inhibitor undergoes base exchange with the nicotinamide moiety of nicotinamide adenine dinucleotide (NAD) to produce the bona fide inhibitor 1AD. We report structures of SARM1 in complex with 1AD, NAD mimetics and the allosteric activator nicotinamide mononucleotide (NMN). NMN binding triggers reorientation of the armadillo repeat (ARM) domains, which disrupts ARM:TIR interactions and leads to formation of a two-stranded TIR domain assembly. The active site spans two molecules in these assemblies, explaining the requirement of TIR domain self-association for NADase activity and axon degeneration. Our results reveal the mechanisms of SARM1 activation and substrate binding, providing rational avenues for the design of new therapeutics targeting SARM1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7nah.cif.gz 7nah.cif.gz | 129.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7nah.ent.gz pdb7nah.ent.gz | 100 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7nah.json.gz 7nah.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/na/7nah https://data.pdbj.org/pub/pdb/validation_reports/na/7nah ftp://data.pdbj.org/pub/pdb/validation_reports/na/7nah ftp://data.pdbj.org/pub/pdb/validation_reports/na/7nah | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7nagC  7naiC  7najC  7nakC  7nalC  6o0rS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||
| 2 | 
| ||||||||||||
| Unit cell |
| ||||||||||||
| Components on special symmetry positions |
|
- Components
Components
| #1: Protein | Mass: 16333.703 Da / Num. of mol.: 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)  Homo sapiens (human) / Gene: SARM1, KIAA0524, SAMD2, SARM / Production host: Homo sapiens (human) / Gene: SARM1, KIAA0524, SAMD2, SARM / Production host:  #2: Chemical | #3: Water | ChemComp-HOH / | Has ligand of interest | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.53 Å3/Da / Density % sol: 51.42 % |
|---|---|
| Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop Details: 0.1 M Bis-Tris propane pH 7.0, 0.2 M potassium thiocyanate, and 10% PEG3350 |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  Australian Synchrotron Australian Synchrotron  / Beamline: MX2 / Wavelength: 0.9537 Å / Beamline: MX2 / Wavelength: 0.9537 Å |
| Detector | Type: DECTRIS EIGER X 16M / Detector: PIXEL / Date: Nov 7, 2020 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9537 Å / Relative weight: 1 |
| Reflection | Resolution: 1.79→48.29 Å / Num. obs: 32174 / % possible obs: 99.9 % / Redundancy: 6.7 % / Biso Wilson estimate: 17.89 Å2 / CC1/2: 0.999 / Rmerge(I) obs: 0.074 / Rpim(I) all: 0.031 / Rrim(I) all: 0.08 / Net I/σ(I): 12.8 |
| Reflection shell | Resolution: 1.79→1.83 Å / Rmerge(I) obs: 0.413 / Mean I/σ(I) obs: 3.1 / Num. unique obs: 1854 / CC1/2: 0.944 / Rpim(I) all: 0.169 / Rrim(I) all: 0.446 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 6O0R Resolution: 1.79→43.12 Å / SU ML: 0.2017 / Cross valid method: FREE R-VALUE / σ(F): 1.35 / Phase error: 17.2791 Stereochemistry target values: GeoStd + Monomer Library + CDL v1.2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 20.94 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.79→43.12 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller






 PDBj
PDBj