[English] 日本語
 Yorodumi
Yorodumi- EMDB-23952: Cryo-EM structure of RecBCD with docked RecBNuc and flexible RecD -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23952 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
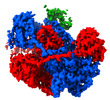
| Title | Cryo-EM structure of RecBCD with docked RecBNuc and flexible RecD | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SF1 helicase / complex / DNA repair / motor protein / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationexodeoxyribonuclease V / exodeoxyribonuclease V activity / exodeoxyribonuclease V complex / clearance of foreign intracellular DNA / DNA translocase activity / DNA 5'-3' helicase / single-stranded DNA helicase activity / recombinational repair / 3'-5' DNA helicase activity / DNA 3'-5' helicase ...exodeoxyribonuclease V / exodeoxyribonuclease V activity / exodeoxyribonuclease V complex / clearance of foreign intracellular DNA / DNA translocase activity / DNA 5'-3' helicase / single-stranded DNA helicase activity / recombinational repair / 3'-5' DNA helicase activity / DNA 3'-5' helicase / ATP-dependent activity, acting on DNA / DNA helicase activity / DNA endonuclease activity / response to radiation / helicase activity / double-strand break repair via homologous recombination / DNA recombination / 5'-3' DNA helicase activity / DNA damage response / magnesium ion binding / ATP hydrolysis activity / DNA binding / ATP binding / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Hao L / Zhang R | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2021 Journal: J Mol Biol / Year: 2021Title: Heterogeneity in E. coli RecBCD Helicase-DNA Binding and Base Pair Melting. Authors: Linxuan Hao / Rui Zhang / Timothy M Lohman /  Abstract: E. coli RecBCD, a helicase/nuclease involved in double stranded (ds) DNA break repair, binds to a dsDNA end and melts out several DNA base pairs (bp) using only its binding free energy. We examined ...E. coli RecBCD, a helicase/nuclease involved in double stranded (ds) DNA break repair, binds to a dsDNA end and melts out several DNA base pairs (bp) using only its binding free energy. We examined RecBCD-DNA initiation complexes using thermodynamic and structural approaches. Measurements of enthalpy changes for RecBCD binding to DNA ends possessing pre-melted ssDNA tails of increasing length suggest that RecBCD interacts with ssDNA as long as 17-18 nucleotides and can melt at least 10-11 bp upon binding a blunt DNA end. Cryo-EM structures of RecBCD alone and in complex with a blunt-ended dsDNA show significant conformational heterogeneities associated with the RecB nuclease domain (RecB) and the RecD subunit. In the absence of DNA, 56% of RecBCD molecules show no density for the RecB nuclease domain, RecB, and all RecBCD molecules show only partial density for RecD. DNA binding reduces these conformational heterogeneities, with 63% of the molecules showing density for both RecD and RecB. This suggests that the RecB domain is dynamic and influenced by DNA binding. The major RecBCD-DNA structural class in which RecB is docked onto RecC shows melting of at least 11 bp from a blunt DNA end, much larger than previously observed. A second structural class in which RecB is not docked shows only four bp melted suggesting that RecBCD complexes transition between states with different extents of DNA melting and that the extent of melting regulates initiation of helicase activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23952.map.gz emd_23952.map.gz | 7.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23952-v30.xml emd-23952-v30.xml emd-23952.xml emd-23952.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23952_fsc.xml emd_23952_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_23952.png emd_23952.png | 170.8 KB | ||
| Filedesc metadata |  emd-23952.cif.gz emd-23952.cif.gz | 7.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23952 http://ftp.pdbj.org/pub/emdb/structures/EMD-23952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23952 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23952 | HTTPS FTP |
-Related structure data
| Related structure data |  7mr0MC  7mr1C  7mr2C  7mr3C  7mr4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23952.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23952.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Heterotrimeric complex of RecBCD
| Entire | Name: Heterotrimeric complex of RecBCD |
|---|---|
| Components |
|
-Supramolecule #1: Heterotrimeric complex of RecBCD
| Supramolecule | Name: Heterotrimeric complex of RecBCD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: RecBCD enzyme subunit RecB
| Macromolecule | Name: RecBCD enzyme subunit RecB / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: exodeoxyribonuclease V |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134.110641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSDVAETLDP LRLPLQGERL IEASAGTGKT FTIAALYLRL LLGLGGSAAF PRPLTVEELL VVTFTEAATA ELRGRIRSNI HELRIACLR ETTDNPLYER LLEEIDDKAQ AAQWLLLAER QMDEAAVFTI HGFCQRMLNL NAFESGMLFE QQLIEDESLL R YQACADFW ...String: MSDVAETLDP LRLPLQGERL IEASAGTGKT FTIAALYLRL LLGLGGSAAF PRPLTVEELL VVTFTEAATA ELRGRIRSNI HELRIACLR ETTDNPLYER LLEEIDDKAQ AAQWLLLAER QMDEAAVFTI HGFCQRMLNL NAFESGMLFE QQLIEDESLL R YQACADFW RRHCYPLPRE IAQVVFETWK GPQALLRDIN RYLQGEAPVI KAPPPDDETL ASRHAQIVAR IDTVKQQWRD AV GELDALI ESSGIDRRKF NRSNQAKWID KISAWAEEET NSYQLPESLE KFSQRFLEDR TKAGGETPRH PLFEAIDQLL AEP LSIRDL VITRALAEIR ETVAREKRRR GELGFDDMLS RLDSALRSES GEVLAAAIRT RFPVAMIDEF QDTDPQQYRI FRRI WHHQP ETALLLIGDP KQAIYAFRGA DIFTYMKARS EVHAHYTLDT NWRSAPGMVN SVNKLFSQTD DAFMFREIPF IPVKS AGKN QALRFVFKGE TQPAMKMWLM EGESCGVGDY QSTMAQVCAA QIRDWLQAGQ RGEALLMNGD DARPVRASDI SVLVRS RQE AAQVRDALTL LEIPSVYLSN RDSVFETLEA QEMLWLLQAV MTPERENTLR SALATSMMGL NALDIETLNN DEHAWDV VV EEFDGYRQIW RKRGVMPMLR ALMSARNIAE NLLATAGGER RLTDILHISE LLQEAGTQLE SEHALVRWLS QHILEPDS N ASSQQMRLES DKHLVQIVTI HKSKGLEYPL VWLPFITNFR VQEQAFYHDR HSFEAVLDLN AAPESVDLAE AERLAEDLR LLYVALTRSV WHCSLGVAPL VRRRGDKKGD TDVHQSALGR LLQKGEPQDA AGLRTCIEAL CDDDIAWQTA QTGDNQPWQV NDVSTAELN AKTLQRLPGD NWRVTSYSGL QQRGHGIAQD LMPRLDVDAA GVASVVEEPT LTPHQFPRGA SPGTFLHSLF E DLDFTQPV DPNWVREKLE LGGFESQWEP VLTEWITAVL QAPLNETGVS LSQLSARNKQ VEMEFYLPIS EPLIASQLDT LI RQFDPLS AGCPPLEFMQ VRGMLKGFID LVFRHEGRYY LLDYKSNWLG EDSSAYTQQA MAAAMQAHRY DLQYQLYTLA LHR YLRHRI ADYDYEHHFG GVIYLFLRGV DKEHPQQGIY TTRPNAGLIA LMDEMFAGMT LEEA UniProtKB: RecBCD enzyme subunit RecB |
-Macromolecule #2: RecBCD enzyme subunit RecC
| Macromolecule | Name: RecBCD enzyme subunit RecC / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: exodeoxyribonuclease V |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 128.974102 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLRVYHSNRL DVLEALMEFI VERERLDDPF EPEMILVQST GMAQWLQMTL SQKFGIAANI DFPLPASFIW DMFVRVLPEI PKESAFNKQ SMSWKLMTLL PQLLEREDFT LLRHYLTDDS DKRKLFQLSS KAADLFDQYL VYRPDWLAQW ETGHLVEGLG E AQAWQAPL ...String: MLRVYHSNRL DVLEALMEFI VERERLDDPF EPEMILVQST GMAQWLQMTL SQKFGIAANI DFPLPASFIW DMFVRVLPEI PKESAFNKQ SMSWKLMTLL PQLLEREDFT LLRHYLTDDS DKRKLFQLSS KAADLFDQYL VYRPDWLAQW ETGHLVEGLG E AQAWQAPL WKALVEYTHQ LGQPRWHRAN LYQRFIETLE SATTCPPGLP SRVFICGISA LPPVYLQALQ ALGKHIEIHL LF TNPCRYY WGDIKDPAYL AKLLTRQRRH SFEDRELPLF RDSENAGQLF NSDGEQDVGN PLLASWGKLG RDYIYLLSDL ESS QELDAF VDVTPDNLLH NIQSDILELE NRAVAGVNIE EFSRSDNKRP LDPLDSSITF HVCHSPQREV EVLHDRLLAM LEED PTLTP RDIIVMVADI DSYSPFIQAV FGSAPADRYL PYAISDRRAR QSHPVLEAFI SLLSLPDSRF VSEDVLALLD VPVLA ARFD ITEEGLRYLR QWVNESGIRW GIDDDNVREL ELPATGQHTW RFGLTRMLLG YAMESAQGEW QSVLPYDESS GLIAEL VGH LASLLMQLNI WRRGLAQERP LEEWLPVCRD MLNAFFLPDA ETEAAMTLIE QQWQAIIAEG LGAQYGDAVP LSLLRDE LA QRLDQERISQ RFLAGPVNIC TLMPMRSIPF KVVCLLGMND GVYPRQLAPL GFDLMSQKPK RGDRSRRDDD RYLFLEAL I SAQQKLYISY IGRSIQDNSE RFPSVLVQEL IDYIGQSHYL PGDEALNCDE SEARVKAHLT CLHTRMPFDP QNYQPGERQ SYAREWLPAA SQAGKAHSEF VQPLPFTLPE TVPLETLQRF WAHPVRAFFQ MRLQVNFRTE DSEIPDTEPF ILEGLSRYQI NQQLLNALV EQDDAERLFR RFRAAGDLPY GAFGEIFWET QCQEMQQLAD RVIACRQPGQ SMEIDLACNG VQITGWLPQV Q PDGLLRWR PSLLSVAQGM QLWLEHLVYC ASGGNGESRL FLRKDGEWRF PPLAAEQALH YLSQLIEGYR EGMSAPLLVL PE SGGAWLK TCYDAQNDAM LDDDSTLQKA RTKFLQAYEG NMMVRGEGDD IWYQRLWRQL TPETMEAIVE QSQRFLLPLF RFN QS UniProtKB: RecBCD enzyme subunit RecC |
-Macromolecule #3: RecBCD enzyme subunit RecD
| Macromolecule | Name: RecBCD enzyme subunit RecD / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: exodeoxyribonuclease V |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.990367 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKLQKQLLEA VEHKQLRPLD VQFALTVAGD EHPAVTLAAA LLSHDAGEGH VCLPLSRLEN NEASHPLLAT CVSEIGELQN WEECLLASQ AVSRGDEPTP MILCGDRLYL NRMWCNERTV ARFFNEVNHA IEVDEALLAQ TLDKLFPVSD EINWQKVAAA V ALTRRISV ...String: MKLQKQLLEA VEHKQLRPLD VQFALTVAGD EHPAVTLAAA LLSHDAGEGH VCLPLSRLEN NEASHPLLAT CVSEIGELQN WEECLLASQ AVSRGDEPTP MILCGDRLYL NRMWCNERTV ARFFNEVNHA IEVDEALLAQ TLDKLFPVSD EINWQKVAAA V ALTRRISV ISGGPGTGKT TTVAKLLAAL IQMADGERCR IRLAAPTGKA AARLTESLGK ALRQLPLTDE QKKRIPEDAS TL HRLLGAQ PGSQRLRHHA GNPLHLDVLV VDEASMIDLP MMSRLIDALP DHARVIFLGD RDQLASVEAG AVLGDICAYA NAG FTAERA RQLSRLTGTH VPAGTGTEAA SLRDSLCLLQ KSYRFGSDSG IGQLAAAINR GDKTAVKTVF QQDFTDIEKR LLQS GEDYI AMLEEALAGY GRYLDLLQAR AEPDLIIQAF NEYQLLCALR EGPFGVAGLN ERIEQFMQQK RKIHRHPHSR WYEGR PVMI ARNDSALGLF NGDIGIALDR GQGTRVWFAM PDGNIKSVQP SRLPEHETTW AMTVHKSQGS EFDHAALILP SQRTPV VTR ELVYTAVTRA RRRLSLYADE RILSAAIATR TERRSGLAAL FSSRE UniProtKB: RecBCD enzyme subunit RecD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 49.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)