[English] 日本語
 Yorodumi
Yorodumi- EMDB-23801: BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibodies Rh... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23801 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibodies Rh.33172 mAb.1 and RM19R | |||||||||
 Map data Map data | 3D map of BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibody Rh.33172 mAb.1 (as Fab fragment). The main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Immune complex / monoclonal antibodies / HIV-1 / BG505 SOSIP / VIRAL PROTEIN | |||||||||
| Biological species |    Human immunodeficiency virus Human immunodeficiency virus | |||||||||
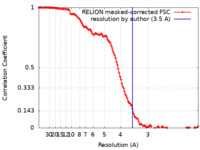
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Antanasijevic A / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: From structure to sequence: Antibody discovery using cryoEM. Authors: Aleksandar Antanasijevic / Charles A Bowman / Robert N Kirchdoerfer / Christopher A Cottrell / Gabriel Ozorowski / Amit A Upadhyay / Kimberly M Cirelli / Diane G Carnathan / Chiamaka A ...Authors: Aleksandar Antanasijevic / Charles A Bowman / Robert N Kirchdoerfer / Christopher A Cottrell / Gabriel Ozorowski / Amit A Upadhyay / Kimberly M Cirelli / Diane G Carnathan / Chiamaka A Enemuo / Leigh M Sewall / Bartek Nogal / Fangzhu Zhao / Bettina Groschel / William R Schief / Devin Sok / Guido Silvestri / Shane Crotty / Steven E Bosinger / Andrew B Ward /  Abstract: One of the rate-limiting steps in analyzing immune responses to vaccines or infections is the isolation and characterization of monoclonal antibodies. Here, we present a hybrid structural and ...One of the rate-limiting steps in analyzing immune responses to vaccines or infections is the isolation and characterization of monoclonal antibodies. Here, we present a hybrid structural and bioinformatic approach to directly assign the heavy and light chains, identify complementarity-determining regions, and discover sequences from cryoEM density maps of serum-derived polyclonal antibodies bound to an antigen. When combined with next-generation sequencing of immune repertoires, we were able to specifically identify clonal family members, synthesize the monoclonal antibodies, and confirm that they interact with the antigen in a manner equivalent to the corresponding polyclonal antibodies. This structure-based approach for identification of monoclonal antibodies from polyclonal sera opens new avenues for analysis of immune responses and iterative vaccine design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23801.map.gz emd_23801.map.gz | 166.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23801-v30.xml emd-23801-v30.xml emd-23801.xml emd-23801.xml | 26.3 KB 26.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23801_fsc.xml emd_23801_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_23801.png emd_23801.png | 122.7 KB | ||
| Masks |  emd_23801_msk_1.map emd_23801_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23801.cif.gz emd-23801.cif.gz | 7.5 KB | ||
| Others |  emd_23801_half_map_1.map.gz emd_23801_half_map_1.map.gz emd_23801_half_map_2.map.gz emd_23801_half_map_2.map.gz | 140.6 MB 140.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23801 http://ftp.pdbj.org/pub/emdb/structures/EMD-23801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23801 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23801 | HTTPS FTP |
-Related structure data
| Related structure data |  7mepMC  7mdtC  7mduC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23801.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23801.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibody Rh.33172 mAb.1 (as Fab fragment). The main map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23801_msk_1.map emd_23801_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 3D map of BG505 SOSIP.v5.2(7S) in complex with...
| File | emd_23801_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibody Rh.33172 mAb.1 (as Fab fragment). Half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: 3D map of BG505 SOSIP.v5.2(7S) in complex with...
| File | emd_23801_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D map of BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibody Rh.33172 mAb.1 (as Fab fragment). Half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibodies Rh...
| Entire | Name: BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibodies Rh.33172 mAb.1 and RM19R |
|---|---|
| Components |
|
-Supramolecule #1: BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibodies Rh...
| Supramolecule | Name: BG505 SOSIP.v5.2(7S) in complex with the monoclonal antibodies Rh.33172 mAb.1 and RM19R type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 Details: The complexes were created by combining BG505 SOSIP.v5.2(7S) and the two antibodies (as Fab fragments) and subsequent SEC purification. |
|---|
-Supramolecule #2: monoclonal antibodies Rh.33172 mAb.1 and RM19R
| Supramolecule | Name: monoclonal antibodies Rh.33172 mAb.1 and RM19R / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2, #4-#5 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: BG505 SOSIP.v5.2(7S)
| Supramolecule | Name: BG505 SOSIP.v5.2(7S) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus Human immunodeficiency virus |
-Macromolecule #1: RM19R mAb Light chain
| Macromolecule | Name: RM19R mAb Light chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.808134 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AIRMTQSPAI LSLSPGERAT LSCRASQSVD SRLAWYQQKP GQSPRLLIYD VSSRATGIPD RFSGSGSGTE FTLTISSLEP EDVAVYFCH QENDWPWTFG QGTKVEIK |
-Macromolecule #2: RM19R mAb Heavy chain
| Macromolecule | Name: RM19R mAb Heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.268667 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVESGPG LVRPSETLSL TCAVSGDSIS TNNGWSWIRQ TPGKGLEWIG YINGRSGSTR YNPSLQSRVT ISTDTSGNQF SLKVNSVTA ADTAKYYCAF FWSTYYKRFD VWGPGVRVTV SS |
-Macromolecule #3: BG505 SOSIPv5.2(7S) - gp120
| Macromolecule | Name: BG505 SOSIPv5.2(7S) - gp120 / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus Human immunodeficiency virus |
| Molecular weight | Theoretical: 74.987219 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKRGLCCVLL LCGAVFVSPS QEIHARFRRG ARAENLWVTV YYGVPVWKDA ETTLFCASDA KAYETKKHNV WATHCCVPTD PNPQEIHLE NVTEEFNMWK NNMVEQMHTD IISLWDQSLK PCVKLTPLCV TLQCTNVTNN ITDDMRGELK NCSFNMTTEL R DKKQKVYS ...String: MKRGLCCVLL LCGAVFVSPS QEIHARFRRG ARAENLWVTV YYGVPVWKDA ETTLFCASDA KAYETKKHNV WATHCCVPTD PNPQEIHLE NVTEEFNMWK NNMVEQMHTD IISLWDQSLK PCVKLTPLCV TLQCTNVTNN ITDDMRGELK NCSFNMTTEL R DKKQKVYS LFYRLDVVQI NENQGNRSNN SNKEYRLINC NTSAITQACP KVSFEPIPIH YCAPAGFAIL KCKDKKFNGT GP CTNVSTV QCTHGIKPVV STQLLLNGSL AEEEVIIRSE NITNNAKNIL VQLNESVQIN CTRPNNNTVK SIRIGPGQWF YYT GDIIGD IRQAHCNVSK ATWNETLGKV VKQLRKHFGN NTIIRFANSS GGDLEVTTHS FNCGGEFFYC NTSGLFNSTW ISNT SVQGS NSTGSNDSIT LPCRIKQIIN MWQRIGQAMY APPIQGVIRC VSNITGLILT RDGGSTNSTT ETFRPGGGDM RDNWR SELY KYKVVKIEPL GVAPTRCKRR VVGRRRRRRA VGIGAVSLGF LGAAGSTMGA ASMTLTVQAR NLLSGIVQQQ SNLLRA PEC QQHLLKDTHW GIKQLQARVL AVEHYLRDQQ LLGIWGCSGK LICCTNVPWN SSWSNRNLSE IWDNMTWLQW DKEISNY TQ IIYGLLEESQ NQQEKNEQDL LELD |
-Macromolecule #4: Rh.33172 mAb.1 Heavy chain
| Macromolecule | Name: Rh.33172 mAb.1 Heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.306693 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKMPGTSVKL SCKTSGYTFT SYNINWVRQA PGQALEWMGW INPNNGTTDY AQKFQGRVTM TRDTSTTTAY MQLNSLRSE DTAVYYCARA RGGYEDDDGY HYTGYGLDSW GQGVVVTVSS |
-Macromolecule #5: Rh.33172 mAb.1 Light chain
| Macromolecule | Name: Rh.33172 mAb.1 Light chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.477829 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSASIGDRVT VTCRASQGIN MQLCWYQLKP GKAPTLLIYG TSGLQTGVSS RFSGSGSGTN FTLTISSLQP EDVATYYCQ QDYTTPFTFG PGTKLDIK |
-Macromolecule #8: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 8 / Number of copies: 51 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: TBS, 0.2um filtered | |||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: OTHER | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force: 0 Wait time: 10s Blot time: 3-7s. | |||||||||
| Details | The sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2050 / Average exposure time: 9.5 sec. / Average electron dose: 44.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)