[English] 日本語
 Yorodumi
Yorodumi- EMDB-2357: Structure and conformational variability of the Mycobacterium tub... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2357 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
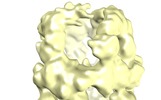
| Title | Structure and conformational variability of the Mycobacterium tuberculosis fatty acid synthase multienzyme complex | |||||||||
 Map data Map data | M. tuberculosis type-I fatty acid synthase - map 1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | M. tuberculosis / fatty acid synthesis / sample heterogeneity / protein flexibility / codimensional principal component analysis | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 17.5 Å | |||||||||
 Authors Authors | Ciccarelli L / Connell SR / Enderle M / Mills DJ / Vonck J / Grininger M | |||||||||
 Citation Citation |  Journal: Structure / Year: 2013 Journal: Structure / Year: 2013Title: Structure and conformational variability of the mycobacterium tuberculosis fatty acid synthase multienzyme complex. Authors: Luciano Ciccarelli / Sean R Connell / Mathias Enderle / Deryck J Mills / Janet Vonck / Martin Grininger /  Abstract: Antibiotic therapy in response to Mycobacterium tuberculosis infections targets de novo fatty acid biosynthesis, which is orchestrated by a 1.9 MDa type I fatty acid synthase (FAS). Here, we ...Antibiotic therapy in response to Mycobacterium tuberculosis infections targets de novo fatty acid biosynthesis, which is orchestrated by a 1.9 MDa type I fatty acid synthase (FAS). Here, we characterize M. tuberculosis FAS by single-particle cryo-electron microscopy and interpret the data by docking the molecular models of yeast and Mycobacterium smegmatis FAS. Our analysis reveals a porous barrel-like structure of considerable conformational variability that is illustrated by the identification of several conformational states with altered topology in the multienzymatic assembly. This demonstrates that the barrel-like structure of M. tuberculosis FAS is not just a static scaffold for the catalytic domains, but may play an active role in coordinating fatty acid synthesis. The conception of M. tuberculosis FAS as a highly dynamic assembly of domains revises the view on bacterial type I fatty acid synthesis and might inspire new strategies for inhibition of de novo fatty acid synthesis in M. tuberculosis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2357.map.gz emd_2357.map.gz | 26.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2357-v30.xml emd-2357-v30.xml emd-2357.xml emd-2357.xml | 8.3 KB 8.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_2357.png emd_2357.png | 2.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2357 http://ftp.pdbj.org/pub/emdb/structures/EMD-2357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2357 | HTTPS FTP |
-Related structure data
| Related structure data |  4v8wMC  2358C  2359C  4v8vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2357.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2357.map.gz / Format: CCP4 / Size: 29.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | M. tuberculosis type-I fatty acid synthase - map 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.28 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : M. tuberculosis type-I fatty acid synthase
| Entire | Name: M. tuberculosis type-I fatty acid synthase |
|---|---|
| Components |
|
-Supramolecule #1000: M. tuberculosis type-I fatty acid synthase
| Supramolecule | Name: M. tuberculosis type-I fatty acid synthase / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 2.0 MDa |
-Macromolecule #1: Type-I fatty acid synthase
| Macromolecule | Name: Type-I fatty acid synthase / type: protein_or_peptide / ID: 1 / Name.synonym: FAS / Oligomeric state: Homohexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 2.0 MDa / Theoretical: 2.0 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Grid | Details: Quantifoils copper grid |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Date | Dec 16, 2010 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 17.5 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: EMAN2, SPARX / Number images used: 9136 |
|---|
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















