+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23013 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human mitochondrial LONP1 in complex with Bortezomib | |||||||||||||||
 Map data Map data | Composite stitched map of human LONP1 in complex with Bortezomib | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | AAA+ / ATPase / protease / mitochondrial / LONP1 / LON / HYDROLASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationoxidation-dependent protein catabolic process / response to aluminum ion / PH domain binding / endopeptidase La / mitochondrial protein catabolic process / mitochondrial DNA metabolic process / G-quadruplex DNA binding / : / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins ...oxidation-dependent protein catabolic process / response to aluminum ion / PH domain binding / endopeptidase La / mitochondrial protein catabolic process / mitochondrial DNA metabolic process / G-quadruplex DNA binding / : / ATP-dependent peptidase activity / protein quality control for misfolded or incompletely synthesized proteins / mitochondrial nucleoid / insulin receptor substrate binding / Mitochondrial unfolded protein response (UPRmt) / chaperone-mediated protein complex assembly / response to hormone / DNA polymerase binding / negative regulation of insulin receptor signaling pathway / Mitochondrial protein degradation / : / mitochondrion organization / protein catabolic process / ADP binding / single-stranded DNA binding / cellular response to oxidative stress / sequence-specific DNA binding / response to hypoxia / single-stranded RNA binding / mitochondrial matrix / serine-type endopeptidase activity / ATP hydrolysis activity / mitochondrion / nucleoplasm / ATP binding / identical protein binding / membrane / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||
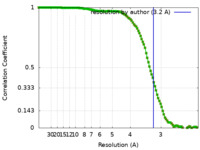
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||||||||
 Authors Authors | Shin M / Watson ER | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Structures of the human LONP1 protease reveal regulatory steps involved in protease activation. Authors: Mia Shin / Edmond R Watson / Albert S Song / Jeffrey T Mindrebo / Scott J Novick / Patrick R Griffin / R Luke Wiseman / Gabriel C Lander /  Abstract: The human mitochondrial AAA+ protein LONP1 is a critical quality control protease involved in regulating diverse aspects of mitochondrial biology including proteostasis, electron transport chain ...The human mitochondrial AAA+ protein LONP1 is a critical quality control protease involved in regulating diverse aspects of mitochondrial biology including proteostasis, electron transport chain activity, and mitochondrial transcription. As such, genetic or aging-associated imbalances in LONP1 activity are implicated in pathologic mitochondrial dysfunction associated with numerous human diseases. Despite this importance, the molecular basis for LONP1-dependent proteolytic activity remains poorly defined. Here, we solved cryo-electron microscopy structures of human LONP1 to reveal the underlying molecular mechanisms governing substrate proteolysis. We show that, like bacterial Lon, human LONP1 adopts both an open and closed spiral staircase orientation dictated by the presence of substrate and nucleotide. Unlike bacterial Lon, human LONP1 contains a second spiral staircase within its ATPase domain that engages substrate as it is translocated toward the proteolytic chamber. Intriguingly, and in contrast to its bacterial ortholog, substrate binding within the central ATPase channel of LONP1 alone is insufficient to induce the activated conformation of the protease domains. To successfully induce the active protease conformation in substrate-bound LONP1, substrate binding within the protease active site is necessary, which we demonstrate by adding bortezomib, a peptidomimetic active site inhibitor of LONP1. These results suggest LONP1 can decouple ATPase and protease activities depending on whether AAA+ or both AAA+ and protease domains bind substrate. Importantly, our structures provide a molecular framework to define the critical importance of LONP1 in regulating mitochondrial proteostasis in health and disease. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23013.map.gz emd_23013.map.gz | 83.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23013-v30.xml emd-23013-v30.xml emd-23013.xml emd-23013.xml | 30.3 KB 30.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23013_fsc.xml emd_23013_fsc.xml emd_23013_fsc_2.xml emd_23013_fsc_2.xml | 13.1 KB 13.1 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_23013.png emd_23013.png | 95.3 KB | ||
| Masks |  emd_23013_msk_1.map emd_23013_msk_1.map emd_23013_msk_2.map emd_23013_msk_2.map | 91.1 MB 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23013.cif.gz emd-23013.cif.gz | 7.9 KB | ||
| Others |  emd_23013_additional_1.map.gz emd_23013_additional_1.map.gz emd_23013_additional_2.map.gz emd_23013_additional_2.map.gz emd_23013_half_map_1.map.gz emd_23013_half_map_1.map.gz emd_23013_half_map_2.map.gz emd_23013_half_map_2.map.gz | 79.5 MB 79.5 MB 71.5 MB 71.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23013 http://ftp.pdbj.org/pub/emdb/structures/EMD-23013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23013 | HTTPS FTP |
-Related structure data
| Related structure data |  7krzMC  7kslC  7ksmC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23013.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23013.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite stitched map of human LONP1 in complex with Bortezomib | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23013_msk_1.map emd_23013_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Mask #2
| File |  emd_23013_msk_2.map emd_23013_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Half-map of protease reconstruction used in composite stitched...
| File | emd_23013_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of protease reconstruction used in composite stitched map of human LONP1 in complex with Bortezomib | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Half-map of protease reconstruction used in composite stitched...
| File | emd_23013_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of protease reconstruction used in composite stitched map of human LONP1 in complex with Bortezomib | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map of ATPase reconstruction used in composite stitched...
| File | emd_23013_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of ATPase reconstruction used in composite stitched map of human LONP1 in complex with Bortezomib | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map of ATPase reconstruction used in composite stitched...
| File | emd_23013_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of ATPase reconstruction used in composite stitched map of human LONP1 in complex with Bortezomib | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human mitochondrial LONP1 in complex with Bortezomib
| Entire | Name: Human mitochondrial LONP1 in complex with Bortezomib |
|---|---|
| Components |
|
-Supramolecule #1: Human mitochondrial LONP1 in complex with Bortezomib
| Supramolecule | Name: Human mitochondrial LONP1 in complex with Bortezomib / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Complexes consisting of homohexameric LONP1 protease from Homo sapiens bound to endogenous co-purified substrate and Bortezomib. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organelle: Mitochondria / Location in cell: Matrix Homo sapiens (human) / Organelle: Mitochondria / Location in cell: Matrix |
| Molecular weight | Theoretical: 462 KDa |
-Macromolecule #1: Lon protease homolog, mitochondrial
| Macromolecule | Name: Lon protease homolog, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: endopeptidase La |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 59.314016 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KDAIEEKFRE RLKELVVPKH VMDVVDEELS KLGLLDNHSS EFNVTRNYLD WLTSIPWGKY SNENLDLARA QAVLEEDHYG MEDVKKRIL EFIAVSQLRG STQGKILCFY GPPGVGKTSI ARSIARALNR EYFRFSVGGM TDVAEIKGHR RTYVGAMPGK I IQCLKKTK ...String: KDAIEEKFRE RLKELVVPKH VMDVVDEELS KLGLLDNHSS EFNVTRNYLD WLTSIPWGKY SNENLDLARA QAVLEEDHYG MEDVKKRIL EFIAVSQLRG STQGKILCFY GPPGVGKTSI ARSIARALNR EYFRFSVGGM TDVAEIKGHR RTYVGAMPGK I IQCLKKTK TENPLILIDE VDKIGRGYQG DPSSALLELL DPEQNANFLD HYLDVPVDLS KVLFICTANV TDTIPEPLRD RM EMINVSG YVAQEKLAIA ERYLVPQARA LCGLDESKAK LSSDVLTLLI KQYCRESGVR NLQKQVEKVL RKSAYKIVSG EAE SVEVTP ENLQDFVGKP VFTVERMYDV TPPGVVMGLA WTAMGGSTLF VETSLRRPQD KDAKGDKDGS LEVTGQLGEV MKES ARIAY TFARAFLMQH APANDYLVTS HIHLHVPEGA TPKDGPSAGC TIVTALLSLA MGRPVRQNLA MTGEVSLTGK ILPVG GIKE KTIAAKRAGV TCIVLPAENK KDFYDLAAFI TEGLEVHFVE HYREIFDIAF UniProtKB: Lon protease homolog, mitochondrial |
-Macromolecule #2: Endogenous co-purified substrate
| Macromolecule | Name: Endogenous co-purified substrate / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.039273 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) |
-Macromolecule #3: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 4 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL...
| Macromolecule | Name: N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE type: ligand / ID: 5 / Number of copies: 6 / Formula: BO2 |
|---|---|
| Molecular weight | Theoretical: 384.237 Da |
| Chemical component information |  ChemComp-BO2: |
-Macromolecule #6: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: Solutions were made fresh from concentrated and filtered using a 0.1 um syringe filter to avoid microbial contamination. Samples were mixed on ice and incubated at 37 degrees C for 30 ...Details: Solutions were made fresh from concentrated and filtered using a 0.1 um syringe filter to avoid microbial contamination. Samples were mixed on ice and incubated at 37 degrees C for 30 minutes to ensure Bortezomib binding. Additional ATP was added to the sample mix on ice 5 minutes prior to vitrification. | |||||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: HOMEMADE PLUNGER Details: 4 uL of sample was applied per grid and manually blotted for 4 seconds followed by immediately plunge-freezing in liquid ethane cooled by liquid nitrogen.. | |||||||||||||||||||||
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 80.0 K / Max: 90.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.14 mrad |
| Details | Coma-free alignment procedure from Herzik & Wu, Nature Methods (2017). Preliminary grid screening was performed manually prior to data collection. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 0-51 / Number grids imaged: 1 / Number real images: 4774 / Average exposure time: 10.4 sec. / Average electron dose: 50.0 e/Å2 Details: Images were collected in counting mode at 5 frames per second. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 1.5 µm / Calibrated defocus min: 0.5 µm / Calibrated magnification: 43478 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Initial homology model was built using SWISS-MODEL and initial rigid body docking was done using UCSF Chimera. |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 52 / Target criteria: Correlation coefficient |
| Output model |  PDB-7krz: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)