[English] 日本語
 Yorodumi
Yorodumi- EMDB-22772: Cryo-EM structure of the Sec complex from S. cerevisiae, wild-typ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22772 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 2 (C2) | |||||||||
 Map data Map data | Unsharpened, lowpass-filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Sec61 / translocon / endoplasmic reticulum / protein translocation / Sec62 / Sec63 / channel / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationmisfolded protein transport / Sec62/Sec63 complex / translocon complex / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / cytosol to endoplasmic reticulum transport / rough endoplasmic reticulum membrane / Ssh1 translocon complex / Sec61 translocon complex / protein-transporting ATPase activity / post-translational protein targeting to endoplasmic reticulum membrane ...misfolded protein transport / Sec62/Sec63 complex / translocon complex / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / cytosol to endoplasmic reticulum transport / rough endoplasmic reticulum membrane / Ssh1 translocon complex / Sec61 translocon complex / protein-transporting ATPase activity / post-translational protein targeting to endoplasmic reticulum membrane / filamentous growth / SRP-dependent cotranslational protein targeting to membrane, translocation / SRP-dependent cotranslational protein targeting to membrane / signal sequence binding / post-translational protein targeting to membrane, translocation / peptide transmembrane transporter activity / nuclear inner membrane / retrograde protein transport, ER to cytosol / protein transmembrane transporter activity / ERAD pathway / guanyl-nucleotide exchange factor activity / cell periphery / ribosome binding / endoplasmic reticulum membrane / structural molecule activity / endoplasmic reticulum / mitochondrion / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Itskanov S / Park E | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2021 Journal: Nat Struct Mol Biol / Year: 2021Title: Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62. Authors: Samuel Itskanov / Katie M Kuo / James C Gumbart / Eunyong Park /  Abstract: Many proteins are transported into the endoplasmic reticulum by the universally conserved Sec61 channel. Post-translational transport requires two additional proteins, Sec62 and Sec63, but their ...Many proteins are transported into the endoplasmic reticulum by the universally conserved Sec61 channel. Post-translational transport requires two additional proteins, Sec62 and Sec63, but their functions are poorly defined. In the present study, we determined cryo-electron microscopy (cryo-EM) structures of several variants of Sec61-Sec62-Sec63 complexes from Saccharomyces cerevisiae and Thermomyces lanuginosus and show that Sec62 and Sec63 induce opening of the Sec61 channel. Without Sec62, the translocation pore of Sec61 remains closed by the plug domain, rendering the channel inactive. We further show that the lateral gate of Sec61 must first be partially opened by interactions between Sec61 and Sec63 in cytosolic and luminal domains, a simultaneous disruption of which completely closes the channel. The structures and molecular dynamics simulations suggest that Sec62 may also prevent lipids from invading the channel through the open lateral gate. Our study shows how Sec63 and Sec62 work together in a hierarchical manner to activate Sec61 for post-translational protein translocation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22772.map.gz emd_22772.map.gz | 32.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22772-v30.xml emd-22772-v30.xml emd-22772.xml emd-22772.xml | 25.3 KB 25.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22772_fsc.xml emd_22772_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_22772.png emd_22772.png | 38.9 KB | ||
| Masks |  emd_22772_msk_1.map emd_22772_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22772.cif.gz emd-22772.cif.gz | 7.1 KB | ||
| Others |  emd_22772_additional_1.map.gz emd_22772_additional_1.map.gz emd_22772_half_map_1.map.gz emd_22772_half_map_1.map.gz emd_22772_half_map_2.map.gz emd_22772_half_map_2.map.gz | 59.5 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22772 http://ftp.pdbj.org/pub/emdb/structures/EMD-22772 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22772 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22772 | HTTPS FTP |
-Validation report
| Summary document |  emd_22772_validation.pdf.gz emd_22772_validation.pdf.gz | 957.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22772_full_validation.pdf.gz emd_22772_full_validation.pdf.gz | 956.8 KB | Display | |
| Data in XML |  emd_22772_validation.xml.gz emd_22772_validation.xml.gz | 16.6 KB | Display | |
| Data in CIF |  emd_22772_validation.cif.gz emd_22772_validation.cif.gz | 21.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22772 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22772 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22772 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22772 | HTTPS FTP |
-Related structure data
| Related structure data |  7kajMC  7kahC  7kaiC  7kakC  7kalC  7kamC  7kanC  7kaoC  7kapC  7kaqC  7karC  7kasC  7katC  7kauC  7kb5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22772.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22772.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened, lowpass-filtered map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.19 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22772_msk_1.map emd_22772_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
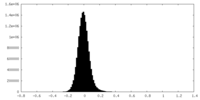
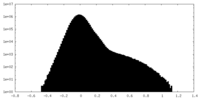
| Density Histograms |
-Additional map: Sharpened map
| File | emd_22772_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_22772_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
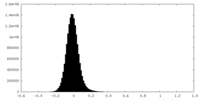
| Density Histograms |
-Half map: #2
| File | emd_22772_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
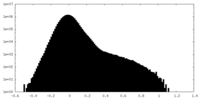
| Density Histograms |
- Sample components
Sample components
-Entire : Endoplasmic reticulum protein-transport machinery Sec complex fro...
| Entire | Name: Endoplasmic reticulum protein-transport machinery Sec complex from yeast |
|---|---|
| Components |
|
-Supramolecule #1: Endoplasmic reticulum protein-transport machinery Sec complex fro...
| Supramolecule | Name: Endoplasmic reticulum protein-transport machinery Sec complex from yeast type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein transport protein SEC61
| Macromolecule | Name: Protein transport protein SEC61 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.978148 KDa |
| Sequence | String: MSSNRVLDLF KPFESFLPEV IAPERKVPYN QKLIWTGVSL LIFLILGQIP LYGIVSSETS DPLYWLRAML ASNRGTLLEL GVSPIITSS MIFQFLQGTQ LLQIRPESKQ DRELFQIAQK VCAIILILGQ ALVVVMTGNY GAPSDLGLPI CLLLIFQLMF A SLIVMLLD ...String: MSSNRVLDLF KPFESFLPEV IAPERKVPYN QKLIWTGVSL LIFLILGQIP LYGIVSSETS DPLYWLRAML ASNRGTLLEL GVSPIITSS MIFQFLQGTQ LLQIRPESKQ DRELFQIAQK VCAIILILGQ ALVVVMTGNY GAPSDLGLPI CLLLIFQLMF A SLIVMLLD ELLSKGYGLG SGISLFTATN IAEQIFWRAF APTTVNSGRG KEFEGAVIAF FHLLAVRKDK KRALVEAFYR TN LPNMFQV LMTVAIFLFV LYLQGFRYEL PIRSTKVRGQ IGIYPIKLFY TSNTPIMLQS ALTSNIFLIS QILFQKYPTN PLI RLIGVW GIRPGTQGPQ MALSGLAYYI QPLMSLSEAL LDPIKTIVYI TFVLGSCAVF SKTWIEISGT SPRDIAKQFK DQGM VINGK RETSIYRELK KIIPTAAAFG GATIGALSVG SDLLGTLGSG ASILMATTTI YGYYEAAAKE GGFTKNLVPG FSDLM UniProtKB: Protein transport protein SEC61 |
-Macromolecule #2: Protein transport protein SSS1
| Macromolecule | Name: Protein transport protein SSS1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.958641 KDa |
| Sequence | String: MARASEKGEE KKQSNNQVEK LVEAPVEFVR EGTQFLAKCK KPDLKEYTKI VKAVGIGFIA VGIIGYAIKL IHIPIRYVIV UniProtKB: Protein transport protein SSS1 |
-Macromolecule #3: Protein transport protein SBH1
| Macromolecule | Name: Protein transport protein SBH1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.723155 KDa |
| Sequence | String: MSSPTPPGGQ RTLQKRKQGS SQKVAASAPK KNTNSNNSIL KIYSDEATGL RVDPLVVLFL AVGFIFSVVA LHVISKVAGK LF UniProtKB: Protein transport protein SBH1 |
-Macromolecule #4: Protein translocation protein SEC63
| Macromolecule | Name: Protein translocation protein SEC63 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 77.93368 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GGSGGSGGSG GSGGSPTNYE YDEASETWPS FILTGLLMVV GPMTLLQIYQ IFFGANAEDG NSGKSKEFNE EVFKNLNEEY TSDEIKQFR RKFDKNSNKK SKIWSRRNII IIVGWILVAI LLQRINSNDA IKDAATKLFD PYEILGISTS ASDRDIKSAY R KLSVKFHP ...String: GGSGGSGGSG GSGGSPTNYE YDEASETWPS FILTGLLMVV GPMTLLQIYQ IFFGANAEDG NSGKSKEFNE EVFKNLNEEY TSDEIKQFR RKFDKNSNKK SKIWSRRNII IIVGWILVAI LLQRINSNDA IKDAATKLFD PYEILGISTS ASDRDIKSAY R KLSVKFHP DKLAKGLTPD EKSVMEETYV QITKAYESLT DELVRQNYLK YGHPDGPQST SHGIALPRFL VDGSASPLLV VC YVALLGL ILPYFVSRWW ARTQSYTKKG IHNVTASNFV SNLVNYKPSE IVTTDLILHW LSFAHEFKQF FPDLQPTDFE KLL QDHINR RDSGKLNNAK FRIVAKCHSL LHGLLDIACG FRNLDIALGA INTFKCIVQA VPLTPNCQIL QLPNVDKEHF ITKT GDIHT LGKLFTLEDA KIGEVLGIKD QAKLNETLRV ASHIPNLKII KADFLVPGEN QVTPSSTPYI SLKVLVRSAK QPLIP TSLI PEENLTEPQD FESQRDPFAM MSKQPLVPYS FAPFFPTKRR GSWCCLVSSQ KDGKILQTPI IIEKLSYKNL NDDKDF FDK RIKMDLTKHE KFDINDWEIG TIKIPLGQPA PETVGDFFFR VIVKSTDYFT TDLDITMNMK VRDSPAVEQV EVYSEED DE YSTDDDETES DDESDASDYT DIDTDTEAED DESPEAGGAT TASGTGENLY FQ UniProtKB: Protein translocation protein SEC63 |
-Macromolecule #5: Translocation protein SEC66
| Macromolecule | Name: Translocation protein SEC66 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.263939 KDa |
| Sequence | String: MSEFNETKFS NNGTFFETEE PIVETKSISV YTPLIYVFIL VVSLVMFASS YRKKQAKKIS EQPSIFDEND AHDLYFQIKE MSENEKIHE KVLKAALLNR GAESVRRSLK LKELAPQINL LYKNGSIGED YWKRFETEVK LIELEFKDTL QEAERLQPGW V QLFVMVCK ...String: MSEFNETKFS NNGTFFETEE PIVETKSISV YTPLIYVFIL VVSLVMFASS YRKKQAKKIS EQPSIFDEND AHDLYFQIKE MSENEKIHE KVLKAALLNR GAESVRRSLK LKELAPQINL LYKNGSIGED YWKRFETEVK LIELEFKDTL QEAERLQPGW V QLFVMVCK EICFNQALSR RYQSILKRKE VCIKEWELKI NNDGRLVN UniProtKB: Translocation protein SEC66 |
-Macromolecule #6: Translocation protein SEC72
| Macromolecule | Name: Translocation protein SEC72 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.63109 KDa |
| Sequence | String: MVTLEYNANS KLITASDAVV ALSTETNIDQ INVLTTSLIG ETNPNFTPQP NEALSKMIKG LFESGMKNLQ QKKLNEALKN VSLAIEMAQ RKRAPWEAFA IQLPELHFML RSKIDLCLIL GKHLEALQDL DFLLGTGLIQ PDVFVRKADC LLKLRQWEEA R ATCERGLA ...String: MVTLEYNANS KLITASDAVV ALSTETNIDQ INVLTTSLIG ETNPNFTPQP NEALSKMIKG LFESGMKNLQ QKKLNEALKN VSLAIEMAQ RKRAPWEAFA IQLPELHFML RSKIDLCLIL GKHLEALQDL DFLLGTGLIQ PDVFVRKADC LLKLRQWEEA R ATCERGLA LAPEDMKLRA LLIETARNLA EYNGE UniProtKB: Translocation protein SEC72 |
-Macromolecule #7: Protein transport protein Sec62
| Macromolecule | Name: Protein transport protein Sec62 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.783888 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 35 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.039 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 49.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 42017 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)