[English] 日本語
 Yorodumi
Yorodumi- EMDB-22617: Cryo-EM structure of heterologous protein complex loaded Thermoto... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22617 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of heterologous protein complex loaded Thermotoga maritima encapsulin capsid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationencapsulin nanocompartment / Hydrolases; Acting on peptide bonds (peptidases) / peptidase activity / iron ion transport / intracellular iron ion homeostasis / proteolysis Similarity search - Function | |||||||||
| Biological species |   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) | |||||||||
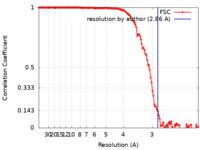
| Method | single particle reconstruction / cryo EM / Resolution: 2.86 Å | |||||||||
 Authors Authors | Xiong X / Sun C / Vago FS / Klose T / Zhu J / Jiang W | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Biomolecules / Year: 2020 Journal: Biomolecules / Year: 2020Title: Cryo-EM Structure of Heterologous Protein Complex Loaded Encapsulin Capsid. Authors: Xiansong Xiong / Chen Sun / Frank S Vago / Thomas Klose / Jiankang Zhu / Wen Jiang /   Abstract: Encapsulin is a class of nanocompartments that is unique in bacteria and archaea to confine enzymatic activities and sequester toxic reaction products. Here we present a 2.87 Å resolution cryo-EM ...Encapsulin is a class of nanocompartments that is unique in bacteria and archaea to confine enzymatic activities and sequester toxic reaction products. Here we present a 2.87 Å resolution cryo-EM structure of encapsulin with heterologous protein complex loaded. It is the first successful case of expressing encapsulin and heterologous cargo protein in the insect cell system. Although we failed to reconstruct the cargo protein complex structure due to the signal interference of the capsid shell, we were able to observe some unique features of the cargo-loaded encapsulin shell, for example, an extra density at the fivefold pore that has not been reported before. These results would lead to a more complete understanding of the encapsulin cargo assembly process of . | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22617.map.gz emd_22617.map.gz | 46 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22617-v30.xml emd-22617-v30.xml emd-22617.xml emd-22617.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22617_fsc.xml emd_22617_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_22617.png emd_22617.png | 265.7 KB | ||
| Others |  emd_22617_half_map_1.map.gz emd_22617_half_map_1.map.gz emd_22617_half_map_2.map.gz emd_22617_half_map_2.map.gz | 45.9 MB 45.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22617 http://ftp.pdbj.org/pub/emdb/structures/EMD-22617 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22617 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22617 | HTTPS FTP |
-Validation report
| Summary document |  emd_22617_validation.pdf.gz emd_22617_validation.pdf.gz | 508.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22617_full_validation.pdf.gz emd_22617_full_validation.pdf.gz | 508.5 KB | Display | |
| Data in XML |  emd_22617_validation.xml.gz emd_22617_validation.xml.gz | 19.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22617 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22617 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22617 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22617 | HTTPS FTP |
-Related structure data
| Related structure data |  7k5wMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22617.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22617.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.072 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
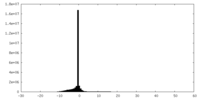
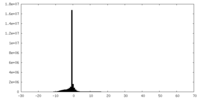
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_22617_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
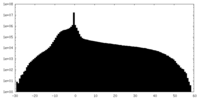
| Density Histograms |
-Half map: #1
| File | emd_22617_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
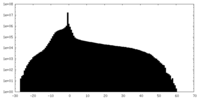
| Density Histograms |
- Sample components
Sample components
-Entire : Thermotoga maritima encapsulin
| Entire | Name: Thermotoga maritima encapsulin |
|---|---|
| Components |
|
-Supramolecule #1: Thermotoga maritima encapsulin
| Supramolecule | Name: Thermotoga maritima encapsulin / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (bacteria) Thermotoga maritima (bacteria) |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 298.15 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.41 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 0.1 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)