[English] 日本語
 Yorodumi
Yorodumi- EMDB-22474: Interphotoreceptor retinoid-binding protein (IRBP) in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22474 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Interphotoreceptor retinoid-binding protein (IRBP) in complex with a monoclonal antibody (F3F5 mAb5) | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | IRBP / retinoid / Interphotoreceptor retinoid-binding protein / Antibody / TRANSPORT PROTEIN-IMMUNE SYSTEM complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcone matrix sheath / interphotoreceptor matrix / all-trans-retinol binding / oleic acid binding / retinal binding / retinol binding / serine-type peptidase activity / proteolysis / extracellular region Similarity search - Function | |||||||||||||||
| Biological species |   | |||||||||||||||
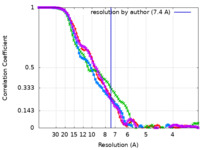
| Method | single particle reconstruction / cryo EM / Resolution: 7.4 Å | |||||||||||||||
 Authors Authors | Sears AE / Albiez S | |||||||||||||||
| Funding support |  United States, United States,  Switzerland, 4 items Switzerland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: FASEB J / Year: 2020 Journal: FASEB J / Year: 2020Title: Single particle cryo-EM of the complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody. Authors: Avery E Sears / Stefan Albiez / Sahil Gulati / Benlian Wang / Philip Kiser / Lubomir Kovacik / Andreas Engel / Henning Stahlberg / Krzysztof Palczewski /   Abstract: Interphotoreceptor retinoid-binding protein (IRBP) is a highly expressed protein secreted by rod and cone photoreceptors that has major roles in photoreceptor homeostasis as well as retinoid and ...Interphotoreceptor retinoid-binding protein (IRBP) is a highly expressed protein secreted by rod and cone photoreceptors that has major roles in photoreceptor homeostasis as well as retinoid and polyunsaturated fatty acid transport between the neural retina and retinal pigment epithelium. Despite two crystal structures reported on fragments of IRBP and decades of research, the overall structure of IRBP and function within the visual cycle remain unsolved. Here, we studied the structure of native bovine IRBP in complex with a monoclonal antibody (mAb5) by cryo-electron microscopy, revealing the tertiary and quaternary structure at sufficient resolution to clearly identify the complex components. Complementary mass spectrometry experiments revealed the structure and locations of N-linked carbohydrate post-translational modifications. This work provides insight into the structure of IRBP, displaying an elongated, flexible three-dimensional architecture not seen among other retinoid-binding proteins. This work is the first step in elucidation of the function of this enigmatic protein. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22474.map.gz emd_22474.map.gz | 2.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22474-v30.xml emd-22474-v30.xml emd-22474.xml emd-22474.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22474_fsc.xml emd_22474_fsc.xml emd_22474_fsc_2.xml emd_22474_fsc_2.xml emd_22474_fsc_3.xml emd_22474_fsc_3.xml emd_22474_fsc_4.xml emd_22474_fsc_4.xml | 8.2 KB 8.3 KB 8.3 KB 8.3 KB | Display Display Display Display |  FSC data file FSC data file |
| Images |  emd_22474.png emd_22474.png | 124.8 KB | ||
| Masks |  emd_22474_msk_1.map emd_22474_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22474.cif.gz emd-22474.cif.gz | 7.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22474 http://ftp.pdbj.org/pub/emdb/structures/EMD-22474 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22474 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22474 | HTTPS FTP |
-Related structure data
| Related structure data |  7jtiMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10388 (Title: Single particle cryo-EM of the complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody EMPIAR-10388 (Title: Single particle cryo-EM of the complex between interphotoreceptor retinoid-binding protein and a monoclonal antibodyData size: 729.9 Data #1: Unaligned multi-frame micrographs of IRBP and antibody [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22474.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22474.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.662 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22474_msk_1.map emd_22474_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between interphotoreceptor retinoid-binding protein and a...
| Entire | Name: Complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody |
|---|---|
| Components |
|
-Supramolecule #1: Complex between interphotoreceptor retinoid-binding protein and a...
| Supramolecule | Name: Complex between interphotoreceptor retinoid-binding protein and a monoclonal antibody type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 193 KDa |
-Supramolecule #2: monoclonal antibody F3F5 mAb5
| Supramolecule | Name: monoclonal antibody F3F5 mAb5 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Interphotoreceptor retinoid-binding protein (IRBP)
| Supramolecule | Name: Interphotoreceptor retinoid-binding protein (IRBP) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#6 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: mAb5 Fab light chain
| Macromolecule | Name: mAb5 Fab light chain / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.596021 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QAVVTQESAL TTSPGETVTL TCRSSTGAVT TSNYANWVQE KPDHLFTGLI GGTNNRAPGV PARFSGSLIG DKAALTITGA QTEDEAIYF CALWYSNHWV FGGGTKLTVL GQPKSSPSVT LFPPSSEELE TNKATLVCTI TDFYPGVVTV DWKVDGTPVT Q GMETTQPS ...String: QAVVTQESAL TTSPGETVTL TCRSSTGAVT TSNYANWVQE KPDHLFTGLI GGTNNRAPGV PARFSGSLIG DKAALTITGA QTEDEAIYF CALWYSNHWV FGGGTKLTVL GQPKSSPSVT LFPPSSEELE TNKATLVCTI TDFYPGVVTV DWKVDGTPVT Q GMETTQPS KQSNNKYMAS SYLTLTARAW ERHSSYSCQV THEGHTVEKS LS |
-Macromolecule #2: mAb5 Fab heavy chain
| Macromolecule | Name: mAb5 Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.777783 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLKESGPG LVAPSQSLSI TCTVSGFLLI SNGVHWVRQP PGKGLEWLGV IWAGGNTNYN SALMSRVSIS KDNSKSQVFL KMKSLQTDD TAMYYCARDF YDYDVFYYAM DYWGQGTSVT VSSAKTTPPS VYPLAPGSAA QTNSMVTLGC LVKGYFPEPV T VTWNSGSL ...String: QVQLKESGPG LVAPSQSLSI TCTVSGFLLI SNGVHWVRQP PGKGLEWLGV IWAGGNTNYN SALMSRVSIS KDNSKSQVFL KMKSLQTDD TAMYYCARDF YDYDVFYYAM DYWGQGTSVT VSSAKTTPPS VYPLAPGSAA QTNSMVTLGC LVKGYFPEPV T VTWNSGSL SSGVHTFPAV LQSDLYTLSS SVTVPSSTWP SETVTCNVAH PASSTKVDKK IVP |
-Macromolecule #3: Retinol-binding protein 3
| Macromolecule | Name: Retinol-binding protein 3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.267301 KDa |
| Sequence | String: VIQRLQEALR EYYTLVDRVP ALLSHLAAMD LSSVVSEDDL VTKLNAGLQA VSEDPRLQVQ VVRPKEASSG PEEEAEEPPE AVPEVPEDE AVRRALVDSV FQVSVLPGNV GYLRFDSFAD ASVLEVLGPY ILHQVWEPLQ DTEHLIMDLR QNPGGPSSAV P LLLSYFQS ...String: VIQRLQEALR EYYTLVDRVP ALLSHLAAMD LSSVVSEDDL VTKLNAGLQA VSEDPRLQVQ VVRPKEASSG PEEEAEEPPE AVPEVPEDE AVRRALVDSV FQVSVLPGNV GYLRFDSFAD ASVLEVLGPY ILHQVWEPLQ DTEHLIMDLR QNPGGPSSAV P LLLSYFQS PDASPVRLFS TYDRRTNITR EHFSQTELLG RPYGTQRGVY LLTSHRTATA AEELAFLMQS LGWATLVGEI TA GSLLHTH TVSLLETPEG GLALTVPVLT FIDNHGECWL GGGVVPDAIV LAEEALDRAQ EVLEFH UniProtKB: Retinol-binding protein 3 |
-Macromolecule #4: Retinol-binding protein 3
| Macromolecule | Name: Retinol-binding protein 3 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.386637 KDa |
| Sequence | String: FSPTLIADMA KIFMDNYCSP EKLTGMEEAI DAASSNTEIL SISDPTMLAN VLTDGVKKTI SDSRVKVTYE PDLILAAPPA MPDIPLEHL AAMIKGTVKV EILEGNIGYL KIQHIIGEEM AQKVGPLLLE YIWDKILPTS AMILDFRSTV TGELSGIPYI V SYFTDPEP ...String: FSPTLIADMA KIFMDNYCSP EKLTGMEEAI DAASSNTEIL SISDPTMLAN VLTDGVKKTI SDSRVKVTYE PDLILAAPPA MPDIPLEHL AAMIKGTVKV EILEGNIGYL KIQHIIGEEM AQKVGPLLLE YIWDKILPTS AMILDFRSTV TGELSGIPYI V SYFTDPEP LIHIDSVYDR TADLTIELWS MPTLLGKRYG TSKPLIILTS KDTLGIAEDV AYCLKNLKRA TIVGENTAGG TV KMSKMKV GDTDFYVTVP VAKSINPITG KSWEINGVAP DVDVAAEDAL DAAIAIIKLR AEIPALAQAA AT |
-Macromolecule #5: Retinol-binding protein 3
| Macromolecule | Name: Retinol-binding protein 3 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.399768 KDa |
| Sequence | String: SLGELVEGTG RLLEAHYARP EVVGQMGALL RAKLAQGAYR TAVDLESLAS QLTADLQEMS GDHRLLVFHS PGEMVAEEAP PPPPVVPSP EELSYLIEAL FKTEVLPGQL GYLRFDAMAE LETVKAVGPQ LVQLVWQKLV DTAALVVDLR YNPGSYSTAV P LLCSYFFE ...String: SLGELVEGTG RLLEAHYARP EVVGQMGALL RAKLAQGAYR TAVDLESLAS QLTADLQEMS GDHRLLVFHS PGEMVAEEAP PPPPVVPSP EELSYLIEAL FKTEVLPGQL GYLRFDAMAE LETVKAVGPQ LVQLVWQKLV DTAALVVDLR YNPGSYSTAV P LLCSYFFE AEPRRHLYSV FDRATSRVTE VWTLPHVTGQ RYGSHKDLYV LVSHTSGSAA EAFAHTMQDL QRATIIGEPT AG GALSVGI YQVGSSALYA SMPTQMAMSA STGEAWDLAG VEPDITVPMS VALSTARDIV TLR UniProtKB: Retinol-binding protein 3 |
-Macromolecule #6: Retinol-binding protein 3
| Macromolecule | Name: Retinol-binding protein 3 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.192508 KDa |
| Sequence | String: PTVLQTAGKL VADNYASPEL GVKMAAELSG LQSRYARVTS EAALAELLQA DLQVLSGDPH LKTAHIPEDA KDRIPGIVPM QIPSPEVFE DLIKFSFHTN VLEGNVGYLR FDMFGDCELL TQVSELLVEH VWKKIVHTDA LIVDMRFNIG GPTSSISALC S YFFDEGPP ...String: PTVLQTAGKL VADNYASPEL GVKMAAELSG LQSRYARVTS EAALAELLQA DLQVLSGDPH LKTAHIPEDA KDRIPGIVPM QIPSPEVFE DLIKFSFHTN VLEGNVGYLR FDMFGDCELL TQVSELLVEH VWKKIVHTDA LIVDMRFNIG GPTSSISALC S YFFDEGPP ILLDKIYNRP NNSVSELWTL SQLEGERYGS KKSMVILTST LTAGAAEEFT YIMKRLGRAL VIGEVTSGGC QP PQTYHVD DTDLYLTIPT ARSVGAADGS SWEGVGVVPD VAVPAEAALT RAQEMLQHT UniProtKB: Retinol-binding protein 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Support film - Film thickness: 1.12 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 3 seconds before plunging.. | |||||||||||||||
| Details | The sample had minor aggregation but was overall monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 1000 pixel / Digitization - Dimensions - Height: 1000 pixel / Number grids imaged: 4 / Number real images: 2649 / Average exposure time: 1.0 sec. / Average electron dose: 80.0 e/Å2 Details: Images were collected in movie mode at 35 frames per second. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: GATAN 910 MULTI-SPECIMEN SINGLE TILT CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 156 / Target criteria: Correlation coefficient |
| Output model |  PDB-7jti: |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)