[English] 日本語
 Yorodumi
Yorodumi- EMDB-22078: Characterization of the SARS-CoV-2 S Protein: Biophysical, Bioche... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22078 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis | |||||||||
 Map data Map data | SARS-CoV2 SPIKE protein reconstruction at 3.22 Angstrom | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | coronavirus / SARS-CoV-2 / SARS-CoV / spike glycoprotein / fusion protein / VIRAL PROTEIN / trimer | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
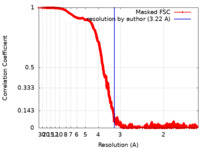
| Method | single particle reconstruction / cryo EM / Resolution: 3.22 Å | |||||||||
 Authors Authors | Herrera NG / Morano NC | |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2020 Journal: bioRxiv / Year: 2020Title: Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis. Authors: Natalia G Herrera / Nicholas C Morano / Alev Celikgil / George I Georgiev / Ryan J Malonis / James H Lee / Karen Tong / Olivia Vergnolle / Aldo B Massimi / Laura Y Yen / Alex J Noble / ...Authors: Natalia G Herrera / Nicholas C Morano / Alev Celikgil / George I Georgiev / Ryan J Malonis / James H Lee / Karen Tong / Olivia Vergnolle / Aldo B Massimi / Laura Y Yen / Alex J Noble / Mykhailo Kopylov / Jeffrey B Bonanno / Sarah C Garrett-Thomson / David B Hayes / Robert H Bortz / Ariel S Wirchnianski / Catalina Florez / Ethan Laudermilch / Denise Haslwanter / J Maximilian Fels / M Eugenia Dieterle / Rohit K Jangra / Jason Barnhill / Amanda Mengotto / Duncan Kimmel / Johanna P Daily / Liise-Anne Pirofski / Kartik Chandran / Michael Brenowitz / Scott J Garforth / Edward T Eng / Jonathan R Lai / Steven C Almo /  Abstract: Coronavirus disease 2019 ( ) is a global health crisis caused by the novel severe acute respiratory syndrome coronavirus 2 ( ), and there is a critical need to produce large quantities of high- ...Coronavirus disease 2019 ( ) is a global health crisis caused by the novel severe acute respiratory syndrome coronavirus 2 ( ), and there is a critical need to produce large quantities of high-quality SARS-CoV-2 Spike ( ) protein for use in both clinical and basic science settings. To address this need, we have evaluated the expression and purification of two previously reported S protein constructs in Expi293F and ExpiCHO-S cells, two different cell lines selected for increased expression of secreted glycoproteins. We show that ExpiCHO-S cells produce enhanced yields of both SARS-CoV-2 S proteins. Biochemical, biophysical, and structural ( ) characterization of the SARS-CoV-2 S proteins produced in both cell lines demonstrate that the reported purification strategy yields high quality S protein (non-aggregated, uniform material with appropriate biochemical and biophysical properties). Importantly, we show that multiple preparations of these two recombinant S proteins from either cell line exhibit identical behavior in two different serology assays. We also evaluate the specificity of S protein-mediated host cell binding by examining interactions with proposed binding partners in the human secretome. In addition, the antigenicity of these proteins is demonstrated by standard ELISAs, and in a flexible protein microarray format. Collectively, we establish an array of metrics for ensuring the production of high-quality S protein to support clinical, biological, biochemical, structural and mechanistic studies to combat the global pandemic caused by SARS-CoV-2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22078.map.gz emd_22078.map.gz | 204 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22078-v30.xml emd-22078-v30.xml emd-22078.xml emd-22078.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22078_fsc.xml emd_22078_fsc.xml | 1 MB | Display |  FSC data file FSC data file |
| Images |  emd_22078.png emd_22078.png | 142.6 KB | ||
| Filedesc metadata |  emd-22078.cif.gz emd-22078.cif.gz | 7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22078 http://ftp.pdbj.org/pub/emdb/structures/EMD-22078 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22078 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22078 | HTTPS FTP |
-Related structure data
| Related structure data |  6x6pMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10433 (Title: Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic Analysis EMPIAR-10433 (Title: Characterization of the SARS-CoV-2 S Protein: Biophysical, Biochemical, Structural, and Antigenic AnalysisData size: 484.3 Data #1: Unaligned multi frame micrographs of SARS CoV-2 SPIKE protein produced in CHO cells [micrographs - multiframe] Data #2: Processed data - aligned frame sum with dose-weighting [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22078.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22078.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SARS-CoV2 SPIKE protein reconstruction at 3.22 Angstrom | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Severe acute respiratory syndrome coronavirus 2
| Entire | Name:  |
|---|---|
| Components |
|
-Supramolecule #1: Severe acute respiratory syndrome coronavirus 2
| Supramolecule | Name: Severe acute respiratory syndrome coronavirus 2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 2697049 Sci species name: Severe acute respiratory syndrome coronavirus 2 Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: Yes / Virus empty: Yes |
|---|
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.824406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CVNLTTRTQL PPAYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPVL PFNDGVYFAS TEKSNIIRG WIFGTTLDSK TQSLLIVNNA TNVVIKVCEF QFCNDPFLGV YYHKNNKSWM ESEFRVYSSA NNCTFEYVSQ P FLMDLEGK ...String: CVNLTTRTQL PPAYTNSFTR GVYYPDKVFR SSVLHSTQDL FLPFFSNVTW FHAIHVSGTN GTKRFDNPVL PFNDGVYFAS TEKSNIIRG WIFGTTLDSK TQSLLIVNNA TNVVIKVCEF QFCNDPFLGV YYHKNNKSWM ESEFRVYSSA NNCTFEYVSQ P FLMDLEGK QGNFKNLREF VFKNIDGYFK IYSKHTPINL VRDLPQGFSA LEPLVDLPIG INITRFQTLL ALHRSYLTPG DS SSGWTAG AAAYYVGYLQ PRTFLLKYNE NGTITDAVDC ALDPLSETKC TLKSFTVEKG IYQTSNFRVQ PTESIVRFPN ITN LCPFGE VFNATRFASV YAWNRKRISN CVADYSVLYN SASFSTFKCY GVSPTKLNDL CFTNVYADSF VIRGDEVRQI APGQ TGKIA DYNYKLPDDF TGCVIAWNSN NLDSKVGGNY NYLYRLFRKS NLKPFERDIS TEIYQAGSTP CNGVEGFNCY FPLQS YGFQ PTNGVGYQPY RVVVLSFELL HAPATVCGPK KSTNLVKNKC VNFNFNGLTG TGVLTESNKK FLPFQQFGRD IADTTD AVR DPQTLEILDI TPCSFGGVSV ITPGTNTSNQ VAVLYQDVNC TEVPVAIHAD QLTPTWRVYS TGSNVFQTRA GCLIGAE HV NNSYECDIPI GAGICASYQT QTNSPGSASS VASQSIIAYT MSLGAENSVA YSNNSIAIPT NFTISVTTEI LPVSMTKT S VDCTMYICGD STECSNLLLQ YGSFCTQLNR ALTGIAVEQD KNTQEVFAQV KQIYKTPPIK DFGGFNFSQI LPDPSKPSK RSFIEDLLFN KVTLADAGFI KQYGDCLGDI AARDLICAQK FNGLTVLPPL LTDEMIAQYT SALLAGTITS GWTFGAGAAL QIPFAMQMA YRFNGIGVTQ NVLYENQKLI ANQFNSAIGK IQDSLSSTAS ALGKLQDVVN QNAQALNTLV KQLSSNFGAI S SVLNDILS RLDPPEAEVQ IDRLITGRLQ SLQTYVTQQL IRAAEIRASA NLAATKMSEC VLGQSKRVDF CGKGYHLMSF PQ SAPHGVV FLHVTYVPAQ EKNFTTAPAI CHDGKAHFPR EGVFVSNGTH WFVTQRNFYE PQIITTDNTF VSGNCDVVIG IVN NTVYDP LQPELDSFKE ELDKYFKNHT SPDVDLGDIS GINASVVNIQ KEIDRLNEVA KNLNESLIDL QELGKYEQGS GYIP EAPRD GQAYVRKDGE WVLLSTFLGR SLEVLFQGPG HHHHHHHHSA WSHPQFEKGG GSGGGGSGGS AWSHPQFEK UniProtKB: Spike glycoprotein |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 33 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 400 / Pretreatment - Type: PLASMA CLEANING | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 1131 / Average exposure time: 2.5 sec. / Average electron dose: 66.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 27-1147 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 120 / Target criteria: CC |
| Output model |  PDB-6x6p: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)