[English] 日本語
 Yorodumi
Yorodumi- EMDB-21477: Escherichia coli transcription-translation complex C6 (TTC-C6) co... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21477 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
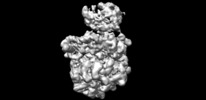
| Title | Escherichia coli transcription-translation complex C6 (TTC-C6) containing mRNA with a 21 nt long spacer, NusA, and fMet-tRNAs at E-site and P-site | |||||||||
 Map data Map data | Escherichia coli transcription-translation complex C6 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial coupled transcription-translation complex / TRANSCRIPTION / RIBOSOME / TRANSCRIPTION-TRANSLATION complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA-templated transcription elongation / positive regulation of ribosome biogenesis / DnaA-L2 complex / negative regulation of DNA-templated DNA replication initiation / DNA-directed RNA polymerase complex / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / regulation of DNA-templated transcription elongation / transcription antitermination / regulation of cell growth ...DNA-templated transcription elongation / positive regulation of ribosome biogenesis / DnaA-L2 complex / negative regulation of DNA-templated DNA replication initiation / DNA-directed RNA polymerase complex / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / regulation of DNA-templated transcription elongation / transcription antitermination / regulation of cell growth / DNA-templated transcription termination / mRNA 5'-UTR binding / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / large ribosomal subunit / transferase activity / ribosome binding / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / protein dimerization activity / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / mRNA binding / DNA-templated transcription / magnesium ion binding / DNA binding / RNA binding / zinc ion binding / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.9 Å | |||||||||
 Authors Authors | Molodtsov V / Wang C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structural basis of transcription-translation coupling. Authors: Chengyuan Wang / Vadim Molodtsov / Emre Firlar / Jason T Kaelber / Gregor Blaha / Min Su / Richard H Ebright /  Abstract: In bacteria, transcription and translation are coupled processes in which the movement of RNA polymerase (RNAP)-synthesizing messenger RNA (mRNA) is coordinated with the movement of the first ...In bacteria, transcription and translation are coupled processes in which the movement of RNA polymerase (RNAP)-synthesizing messenger RNA (mRNA) is coordinated with the movement of the first ribosome-translating mRNA. Coupling is modulated by the transcription factors NusG (which is thought to bridge RNAP and the ribosome) and NusA. Here, we report cryo-electron microscopy structures of transcription-translation complexes (TTCs) containing different-length mRNA spacers between RNAP and the ribosome active-center P site. Structures of TTCs containing short spacers show a state incompatible with NusG bridging and NusA binding (TTC-A, previously termed "expressome"). Structures of TTCs containing longer spacers reveal a new state compatible with NusG bridging and NusA binding (TTC-B) and reveal how NusG bridges and NusA binds. We propose that TTC-B mediates NusG- and NusA-dependent transcription-translation coupling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21477.map.gz emd_21477.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21477-v30.xml emd-21477-v30.xml emd-21477.xml emd-21477.xml | 88.2 KB 88.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_21477.png emd_21477.png | 72.6 KB | ||
| Filedesc metadata |  emd-21477.cif.gz emd-21477.cif.gz | 16.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21477 http://ftp.pdbj.org/pub/emdb/structures/EMD-21477 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21477 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21477 | HTTPS FTP |
-Related structure data
| Related structure data |  6vyzMC  6vu3C  6vyqC  6vyrC  6vysC  6vytC  6vyuC  6vywC  6vyxC  6vyyC  6vz2C  6vz3C  6vz5C  6vz7C  6vzjC  6x6tC  6x7fC  6x7kC  6x9qC  6xdqC  6xdrC  6xgfC  6xiiC  6xijC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21477.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21477.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Escherichia coli transcription-translation complex C6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.9399 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Escherichia coli transcription-translation complex C6 (TTC-C6) co...
+Supramolecule #1: Escherichia coli transcription-translation complex C6 (TTC-C6) co...
+Macromolecule #1: 50S ribosomal protein L21
+Macromolecule #2: 50S ribosomal protein L22
+Macromolecule #3: 50S ribosomal protein L23
+Macromolecule #4: 50S ribosomal protein L24
+Macromolecule #5: 50S ribosomal protein L25
+Macromolecule #10: DNA-directed RNA polymerase subunit beta
+Macromolecule #11: Transcription termination/antitermination protein NusG
+Macromolecule #12: DNA-directed RNA polymerase subunit alpha
+Macromolecule #13: DNA-directed RNA polymerase subunit beta'
+Macromolecule #14: DNA-directed RNA polymerase subunit omega
+Macromolecule #15: 30S ribosomal protein S18
+Macromolecule #17: 30S ribosomal protein S20
+Macromolecule #18: 30S ribosomal protein S21
+Macromolecule #19: 30S ribosomal protein S2
+Macromolecule #20: 30S ribosomal protein S1
+Macromolecule #21: 30S ribosomal protein S3
+Macromolecule #22: 30S ribosomal protein S4
+Macromolecule #23: 30S ribosomal protein S5
+Macromolecule #24: 30S ribosomal protein S6
+Macromolecule #25: 30S ribosomal protein S7
+Macromolecule #26: 30S ribosomal protein S8
+Macromolecule #27: 30S ribosomal protein S9
+Macromolecule #28: 30S ribosomal protein S10
+Macromolecule #29: 30S ribosomal protein S11
+Macromolecule #30: 30S ribosomal protein S12
+Macromolecule #31: 30S ribosomal protein S14
+Macromolecule #32: 30S ribosomal protein S15
+Macromolecule #33: 30S ribosomal protein S16
+Macromolecule #34: 30S ribosomal protein S17
+Macromolecule #35: 30S ribosomal protein S19
+Macromolecule #36: 30S ribosomal protein S13
+Macromolecule #39: 50S ribosomal protein L27
+Macromolecule #40: 50S ribosomal protein L28
+Macromolecule #42: 50S ribosomal protein L29
+Macromolecule #43: 50S ribosomal protein L30
+Macromolecule #44: 50S ribosomal protein L31
+Macromolecule #45: 50S ribosomal protein L2
+Macromolecule #46: 50S ribosomal protein L32
+Macromolecule #47: 50S ribosomal protein L3
+Macromolecule #48: 50S ribosomal protein L33
+Macromolecule #49: 50S ribosomal protein L4
+Macromolecule #50: 50S ribosomal protein L34
+Macromolecule #51: 50S ribosomal protein L5
+Macromolecule #52: 50S ribosomal protein L35
+Macromolecule #53: 50S ribosomal protein L6
+Macromolecule #54: 50S ribosomal protein L36
+Macromolecule #55: 50S ribosomal protein L9
+Macromolecule #56: 50S ribosomal protein L13
+Macromolecule #57: 50S ribosomal protein L14
+Macromolecule #58: 50S ribosomal protein L15
+Macromolecule #59: 50S ribosomal protein L16
+Macromolecule #60: 50S ribosomal protein L17
+Macromolecule #61: 50S ribosomal protein L18
+Macromolecule #62: 50S ribosomal protein L19
+Macromolecule #63: 50S ribosomal protein L20
+Macromolecule #6: NT DNA
+Macromolecule #7: T DNA
+Macromolecule #8: mRNA with 21 nt long spacer
+Macromolecule #9: E-site and P-site tRNA (fMet)
+Macromolecule #16: 16S rRNA
+Macromolecule #37: mRNA in the ribosomal RNA entrance pore
+Macromolecule #38: 23S rRNA
+Macromolecule #41: 5S rRNA
+Macromolecule #64: MAGNESIUM ION
+Macromolecule #65: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 9.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 5913 |
| Initial angle assignment | Type: COMMON LINE |
| Final angle assignment | Type: COMMON LINE |
 Movie
Movie Controller
Controller



































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















