+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21367 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Cas13(crRNA) | |||||||||
 Map data Map data | Cas13(crRNA) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas system / Cas13 / IMMUNE SYSTEM / TypeVI | |||||||||
| Function / homology | : / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / RNA binding / CRISPR-associated endoribonuclease Cas13a Function and homology information Function and homology information | |||||||||
| Biological species |  Listeria seeligeri (bacteria) / Listeria seeligeri (bacteria) /  Listeria seeligeri serovar 1/2b (strain ATCC 35967 / DSM 20751 / CIP 100100 / SLCC 3954) (bacteria) / Listeria seeligeri serovar 1/2b (strain ATCC 35967 / DSM 20751 / CIP 100100 / SLCC 3954) (bacteria) /  Listeria seeligeri serovar 1/2b str. SLCC3954 (bacteria) Listeria seeligeri serovar 1/2b str. SLCC3954 (bacteria) | |||||||||
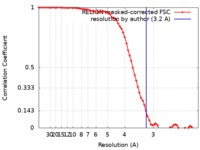
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Jia N / Meeske AJ | |||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity. Authors: Alexander J Meeske / Ning Jia / Alice K Cassel / Albina Kozlova / Jingqiu Liao / Martin Wiedmann / Dinshaw J Patel / Luciano A Marraffini /  Abstract: The CRISPR RNA (crRNA)-guided nuclease Cas13 recognizes complementary viral transcripts to trigger the degradation of both host and viral RNA during the type VI CRISPR-Cas antiviral response. ...The CRISPR RNA (crRNA)-guided nuclease Cas13 recognizes complementary viral transcripts to trigger the degradation of both host and viral RNA during the type VI CRISPR-Cas antiviral response. However, how viruses can counteract this immunity is not known. We describe a listeriaphage (ϕLS46) encoding an anti-CRISPR protein (AcrVIA1) that inactivates the type VI-A CRISPR system of Using genetics, biochemistry, and structural biology, we found that AcrVIA1 interacts with the guide-exposed face of Cas13a, preventing access to the target RNA and the conformational changes required for nuclease activation. Unlike inhibitors of DNA-cleaving Cas nucleases, which cause limited immunosuppression and require multiple infections to bypass CRISPR defenses, a single dose of AcrVIA1 delivered by an individual virion completely dismantles type VI-A CRISPR-mediated immunity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21367.map.gz emd_21367.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21367-v30.xml emd-21367-v30.xml emd-21367.xml emd-21367.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21367_fsc.xml emd_21367_fsc.xml | 7 KB | Display |  FSC data file FSC data file |
| Images |  emd_21367.png emd_21367.png | 71.8 KB | ||
| Filedesc metadata |  emd-21367.cif.gz emd-21367.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21367 http://ftp.pdbj.org/pub/emdb/structures/EMD-21367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21367 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21367 | HTTPS FTP |
-Validation report
| Summary document |  emd_21367_validation.pdf.gz emd_21367_validation.pdf.gz | 390.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21367_full_validation.pdf.gz emd_21367_full_validation.pdf.gz | 390.1 KB | Display | |
| Data in XML |  emd_21367_validation.xml.gz emd_21367_validation.xml.gz | 9.4 KB | Display | |
| Data in CIF |  emd_21367_validation.cif.gz emd_21367_validation.cif.gz | 12.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21367 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21367 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21367 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21367 | HTTPS FTP |
-Related structure data
| Related structure data |  6vrcMC  6vrbC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_21367.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21367.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cas13(crRNA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cas13-crRNA complex
| Entire | Name: Cas13-crRNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Cas13-crRNA complex
| Supramolecule | Name: Cas13-crRNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Listeria seeligeri (bacteria) Listeria seeligeri (bacteria) |
| Molecular weight | Theoretical: 130 KDa |
-Macromolecule #1: CRISPR-associated endoribonuclease Cas13a
| Macromolecule | Name: CRISPR-associated endoribonuclease Cas13a / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Listeria seeligeri serovar 1/2b (strain ATCC 35967 / DSM 20751 / CIP 100100 / SLCC 3954) (bacteria) Listeria seeligeri serovar 1/2b (strain ATCC 35967 / DSM 20751 / CIP 100100 / SLCC 3954) (bacteria)Strain: ATCC 35967 / DSM 20751 / CIP 100100 / SLCC 3954 |
| Molecular weight | Theoretical: 133.295297 KDa |
| Recombinant expression | Organism:  Listeria seeligeri (bacteria) Listeria seeligeri (bacteria) |
| Sequence | String: MWISIKTLIH HLGVLFFCDY MYNRREKKII EVKTMRITKV EVDRKKVLIS RDKNGGKLVY ENEMQDNTEQ IMHHKKSSFY KSVVNKTIC RPEQKQMKKL VHGLLQENSQ EKIKVSDVTK LNISNFLNHR FKKSLYYFPE NSPDKSEEYR IEINLSQLLE D SLKKQQGT ...String: MWISIKTLIH HLGVLFFCDY MYNRREKKII EVKTMRITKV EVDRKKVLIS RDKNGGKLVY ENEMQDNTEQ IMHHKKSSFY KSVVNKTIC RPEQKQMKKL VHGLLQENSQ EKIKVSDVTK LNISNFLNHR FKKSLYYFPE NSPDKSEEYR IEINLSQLLE D SLKKQQGT FICWESFSKD MELYINWAEN YISSKTKLIK KSIRNNRIQS TESRSGQLMD RYMKDILNKN KPFDIQSVSE KY QLEKLTS ALKATFKEAK KNDKEINYKL KSTLQNHERQ IIEELKENSE LNQFNIEIRK HLETYFPIKK TNRKVGDIRN LEI GEIQKI VNHRLKNKIV QRILQEGKLA SYEIESTVNS NSLQKIKIEE AFALKFINAC LFASNNLRNM VYPVCKKDIL MIGE FKNSF KEIKHKKFIR QWSQFFSQEI TVDDIELASW GLRGAIAPIR NEIIHLKKHS WKKFFNNPTF KVKKSKIING KTKDV TSEF LYKETLFKDY FYSELDSVPE LIINKMESSK ILDYYSSDQL NQVFTIPNFE LSLLTSAVPF APSFKRVYLK GFDYQN QDE AQPDYNLKLN IYNEKAFNSE AFQAQYSLFK MVYYQVFLPQ FTTNNDLFKS SVDFILTLNK ERKGYAKAFQ DIRKMNK DE KPSEYMSYIQ SQLMLYQKKQ EEKEKINHFE KFINQVFIKG FNSFIEKNRL TYICHPTKNT VPENDNIEIP FHTDMDDS N IAFWLMCKLL DAKQLSELRN EMIKFSCSLQ STEEISTFTK AREVIGLALL NGEKGCNDWK ELFDDKEAWK KNMSLYVSE ELLQSLPYTQ EDGQTPVINR SIDLVKKYGT ETILEKLFSS SDDYKVSAKD IAKLHEYDVT EKIAQQESLH KQWIEKPGLA RDSAWTKKY QNVINDISNY QWAKTKVELT QVRHLHQLTI DLLSRLAGYM SIADRDFQFS SNYILERENS EYRVTSWILL S ENKNKNKY NDYELYNLKN ASIKVSSKND PQLKVDLKQL RLTLEYLELF DNRLKEKRNN ISHFNYLNGQ LGNSILELFD DA RDVLSYD RKLKNAVSKS LKEILSSHGM EVTFKPLYQT NHHLKIDKLQ PKKIHHLGEK STVSSNQVSN EYCQLVRTLL TMK HHHHHH UniProtKB: CRISPR-associated endoribonuclease Cas13a |
-Macromolecule #2: RNA (51-MER)
| Macromolecule | Name: RNA (51-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Listeria seeligeri serovar 1/2b str. SLCC3954 (bacteria) Listeria seeligeri serovar 1/2b str. SLCC3954 (bacteria) |
| Molecular weight | Theoretical: 19.049383 KDa |
| Sequence | String: GACUACCUCU AUAUGAAAGA GGACUAAAAC CAUAUUUCCA AACUCCACUU UGACUACACC GENBANK: GENBANK: FN557490.1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Formula: Tris / Details: 20 mM Tris, pH 7.5, 150 mM NaCl, 2 mM DTT |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.23 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)