+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20968 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ELIC-propylammonium complex in POPC-only nanodiscs | |||||||||
 Map data Map data | ELIC-propylammonium complex in POPC-only nanodiscs | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Pentameric Ligand-gated Ion Channels / POPC / Propylamonium / Nanodisc / Cys-loop receptor / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationextracellular ligand-gated monoatomic ion channel activity / transmembrane signaling receptor activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |  Dickeya dadantii 3937 (bacteria) / Dickeya dadantii 3937 (bacteria) /  Dickeya dadantii (strain 3937) (bacteria) Dickeya dadantii (strain 3937) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Grosman C / Kumar P | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Cryo-EM structures of a lipid-sensitive pentameric ligand-gated ion channel embedded in a phosphatidylcholine-only bilayer. Authors: Pramod Kumar / Yuhang Wang / Zhening Zhang / Zhiyu Zhao / Gisela D Cymes / Emad Tajkhorshid / Claudio Grosman /  Abstract: The lipid dependence of the nicotinic acetylcholine receptor from the electric organ has long been recognized, and one of the most consistent experimental observations is that, when reconstituted in ...The lipid dependence of the nicotinic acetylcholine receptor from the electric organ has long been recognized, and one of the most consistent experimental observations is that, when reconstituted in membranes formed by zwitterionic phospholipids alone, exposure to agonist fails to elicit ion-flux activity. More recently, it has been suggested that the bacterial homolog ELIC ( ligand-gated ion channel) has a similar lipid sensitivity. As a first step toward the elucidation of the structural basis of this phenomenon, we solved the structures of ELIC embedded in palmitoyl-oleoyl-phosphatidylcholine- (POPC-) only nanodiscs in both the unliganded (4.1-Å resolution) and agonist-bound (3.3 Å) states using single-particle cryoelectron microscopy. Comparison of the two structural models revealed that the largest differences occur at the level of loop C-at the agonist-binding sites-and the loops at the interface between the extracellular and transmembrane domains (ECD and TMD, respectively). On the other hand, the transmembrane pore is occluded in a remarkably similar manner in both structures. A straightforward interpretation of these findings is that POPC-only membranes frustrate the ECD-TMD coupling in such a way that the "conformational wave" of liganded-receptor gating takes place in the ECD and the interfacial M2-M3 linker but fails to penetrate the membrane and propagate into the TMD. Furthermore, analysis of the structural models and molecular simulations suggested that the higher affinity for agonists characteristic of the open- and desensitized-channel conformations results, at least in part, from the tighter confinement of the ligand to its binding site; this limits the ligand's fluctuations, and thus delays its escape into bulk solvent. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20968.map.gz emd_20968.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20968-v30.xml emd-20968-v30.xml emd-20968.xml emd-20968.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20968.png emd_20968.png | 43.5 KB | ||
| Filedesc metadata |  emd-20968.cif.gz emd-20968.cif.gz | 5.7 KB | ||
| Others |  emd_20968_half_map_1.map.gz emd_20968_half_map_1.map.gz emd_20968_half_map_2.map.gz emd_20968_half_map_2.map.gz | 23.4 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20968 http://ftp.pdbj.org/pub/emdb/structures/EMD-20968 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20968 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20968 | HTTPS FTP |
-Validation report
| Summary document |  emd_20968_validation.pdf.gz emd_20968_validation.pdf.gz | 667.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20968_full_validation.pdf.gz emd_20968_full_validation.pdf.gz | 667 KB | Display | |
| Data in XML |  emd_20968_validation.xml.gz emd_20968_validation.xml.gz | 10.9 KB | Display | |
| Data in CIF |  emd_20968_validation.cif.gz emd_20968_validation.cif.gz | 12.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20968 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20968 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20968 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20968 | HTTPS FTP |
-Related structure data
| Related structure data |  6v03MC  6v0bC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20968.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20968.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ELIC-propylammonium complex in POPC-only nanodiscs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0961 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: ELIC-propylammonium complex in POPC-only nanodiscs
| File | emd_20968_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ELIC-propylammonium complex in POPC-only nanodiscs | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: ELIC-propylammonium complex in POPC-only nanodiscs
| File | emd_20968_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ELIC-propylammonium complex in POPC-only nanodiscs | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ELIC-propylammonium complex in POPC-only nanodiscs.
| Entire | Name: ELIC-propylammonium complex in POPC-only nanodiscs. |
|---|---|
| Components |
|
-Supramolecule #1: ELIC-propylammonium complex in POPC-only nanodiscs.
| Supramolecule | Name: ELIC-propylammonium complex in POPC-only nanodiscs. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Dickeya dadantii 3937 (bacteria) Dickeya dadantii 3937 (bacteria) |
-Macromolecule #1: Gamma-aminobutyric-acid receptor subunit beta-1
| Macromolecule | Name: Gamma-aminobutyric-acid receptor subunit beta-1 / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Dickeya dadantii (strain 3937) (bacteria) / Strain: 3937 Dickeya dadantii (strain 3937) (bacteria) / Strain: 3937 |
| Molecular weight | Theoretical: 36.664742 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: APADNAADAR PVDVSVSIFI NKIYGVNTLE QTYKVDGYIV AQWTGKPRKT PGDKPLIVEN TQIERWINNG LWVPALEFIN VVGSPDTGN KRLMLFPDGR VIYNARFLGS FSNDMDFRLF PFDRQQFVLE LEPFSYNNQQ LRFSDIQVYT ENIDNEEIDE W WIRGKAST ...String: APADNAADAR PVDVSVSIFI NKIYGVNTLE QTYKVDGYIV AQWTGKPRKT PGDKPLIVEN TQIERWINNG LWVPALEFIN VVGSPDTGN KRLMLFPDGR VIYNARFLGS FSNDMDFRLF PFDRQQFVLE LEPFSYNNQQ LRFSDIQVYT ENIDNEEIDE W WIRGKAST HISDIRYDHL SSVQPNQNEF SRITVRIDAV RNPSYYLWSF ILPLGLIIAA SWSVFWLESF SERLQTSFTL ML TVVAYAF YTSNILPRLP YTTVIDQMII AGYGSIFAAI LLIIFAHHRQ ANGVEDDLLI QRCRLAFPLG FLAIGCVLVI RGI UniProtKB: Gamma-aminobutyric-acid receptor subunit beta-1 |
-Macromolecule #2: 3-AMINOPROPANE
| Macromolecule | Name: 3-AMINOPROPANE / type: ligand / ID: 2 / Number of copies: 10 / Formula: 3CN |
|---|---|
| Molecular weight | Theoretical: 59.11 Da |
| Chemical component information | 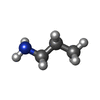 ChemComp-3CN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: 150 NaCl and 10 sodium phosphate, pH 8.0. | ||||||||||||
| Grid | Model: Homemade / Material: GOLD | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: SPOTITON |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: DIRECT ELECTRON DE-16 (4k x 4k) / Detector mode: COUNTING / Average electron dose: 63.75 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6v03: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)