[English] 日本語
 Yorodumi
Yorodumi- EMDB-20556: Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonell... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20556 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonella SPI-1 injectisome NC-base | |||||||||
 Map data Map data | Focussed refinement of the Salmonella SPI-1 injectisome needle complex components PrgHK, SpaPQR, PrgIJ, and InvG-N0N1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacterial secretion / type III secretion system / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationThe IPAF inflammasome / type III protein secretion system complex / type II protein secretion system complex / protein secretion by the type III secretion system / protein secretion / protein targeting / cell outer membrane / protein transport / cell surface / extracellular region ...The IPAF inflammasome / type III protein secretion system complex / type II protein secretion system complex / protein secretion by the type III secretion system / protein secretion / protein targeting / cell outer membrane / protein transport / cell surface / extracellular region / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Hu J / Worrall LJ / Strynadka NCJ / Hong C / Atkinson CE / Yu Z | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2019 Journal: Nat Microbiol / Year: 2019Title: T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly. Authors: Jinhong Hu / Liam J Worrall / Marija Vuckovic / Chuan Hong / Wanyin Deng / Claire E Atkinson / B Brett Finlay / Zhiheng Yu / Natalie C J Strynadka /   Abstract: The bacterial injectisome is a syringe-shaped macromolecular nanomachine utilized by many pathogenic Gram-negative bacteria, including the causative agents of plague, typhoid fever, whooping cough, ...The bacterial injectisome is a syringe-shaped macromolecular nanomachine utilized by many pathogenic Gram-negative bacteria, including the causative agents of plague, typhoid fever, whooping cough, sexually transmitted infections and major nosocomial infections. Bacterial proteins destined for self-assembly and host-cell targeting are translocated by the injectisome in a process known as type III secretion (T3S). The core structure is the ~4 MDa needle complex (NC), built on a foundation of three highly oligomerized ring-forming proteins that create a hollow scaffold spanning the bacterial inner membrane (IM) (24-mer ring-forming proteins PrgH and PrgK in the Salmonella enterica serovar Typhimurium Salmonella pathogenicity island 1 (SPI-1) type III secretion system (T3SS)) and outer membrane (OM) (15-mer InvG, a member of the broadly conserved secretin pore family). An internalized helical needle projects from the NC and bacterium, ultimately forming a continuous passage to the host, for delivery of virulence effectors. Here, we have captured snapshots of the entire prototypical SPI-1 NC in four distinct needle assembly states, including near-atomic resolution, and local reconstructions in the absence and presence of the needle. These structures reveal the precise localization and molecular interactions of the internalized SpaPQR 'export apparatus' complex, which is intimately encapsulated and stabilized within the IM rings in the manner of a nanodisc, and to which the PrgJ rod directly binds and functions as an initiator and anchor of needle polymerization. We also describe the molecular details of the extensive and continuous coupling interface between the OM secretin and IM rings, which is remarkably facilitated by a localized 16-mer stoichiometry in the periplasmic-most coupling domain of the otherwise 15-mer InvG oligomer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20556.map.gz emd_20556.map.gz | 4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20556-v30.xml emd-20556-v30.xml emd-20556.xml emd-20556.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20556.png emd_20556.png | 59.6 KB | ||
| Filedesc metadata |  emd-20556.cif.gz emd-20556.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20556 http://ftp.pdbj.org/pub/emdb/structures/EMD-20556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20556 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20556 | HTTPS FTP |
-Validation report
| Summary document |  emd_20556_validation.pdf.gz emd_20556_validation.pdf.gz | 440.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20556_full_validation.pdf.gz emd_20556_full_validation.pdf.gz | 440 KB | Display | |
| Data in XML |  emd_20556_validation.xml.gz emd_20556_validation.xml.gz | 6 KB | Display | |
| Data in CIF |  emd_20556_validation.cif.gz emd_20556_validation.cif.gz | 6.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20556 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20556 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20556 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20556 | HTTPS FTP |
-Related structure data
| Related structure data |  6q16MC  6peeC  6pemC  6pepC  6q14C  6q15C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20556.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20556.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focussed refinement of the Salmonella SPI-1 injectisome needle complex components PrgHK, SpaPQR, PrgIJ, and InvG-N0N1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.71 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
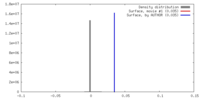
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : InvGN0N1:SpaPQR:PrgHK from SPI-1 injectisome NC-base
| Entire | Name: InvGN0N1:SpaPQR:PrgHK from SPI-1 injectisome NC-base |
|---|---|
| Components |
|
-Supramolecule #1: InvGN0N1:SpaPQR:PrgHK from SPI-1 injectisome NC-base
| Supramolecule | Name: InvGN0N1:SpaPQR:PrgHK from SPI-1 injectisome NC-base / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) |
| Molecular weight | Theoretical: 1.5 MDa |
-Macromolecule #1: Lipoprotein PrgK
| Macromolecule | Name: Lipoprotein PrgK / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 |
| Molecular weight | Theoretical: 28.245287 KDa |
| Sequence | String: MIRRYLYTFL LVMTLAGCKD KDLLKGLDQE QANEVIAVLQ MHNIEANKID SGKLGYSITV AEPDFTAAVY WIKTYQLPPR PRVEIAQMF PADSLVSSPR AEKARLYSAI EQRLEQSLQT MEGVLSARVH ISYDIDAGEN GRPPKPVHLS ALAVYERGSP L AHQISDIK ...String: MIRRYLYTFL LVMTLAGCKD KDLLKGLDQE QANEVIAVLQ MHNIEANKID SGKLGYSITV AEPDFTAAVY WIKTYQLPPR PRVEIAQMF PADSLVSSPR AEKARLYSAI EQRLEQSLQT MEGVLSARVH ISYDIDAGEN GRPPKPVHLS ALAVYERGSP L AHQISDIK RFLKNSFADV DYDNISVVLS ERSDAQLQAP GTPVKRNSFA TSWIVLIILL SVMSAGFGVW YYKNHYARNK KG ITADDKA KSSNE UniProtKB: Lipoprotein PrgK |
-Macromolecule #2: Protein InvG
| Macromolecule | Name: Protein InvG / type: protein_or_peptide / ID: 2 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 |
| Molecular weight | Theoretical: 61.835559 KDa |
| Sequence | String: MKTHILLARV LACAALVLVT PGYSSEKIPV TGSGFVAKDD SLRTFFDAMA LQLKEPVIVS KMAARKKITG NFEFHDPNAL LEKLSLQLG LIWYFDGQAI YIYDASEMRN AVVSLRNVSL NEFNNFLKRS GLYNKNYPLR GDNRKGTFYV SGPPVYVDMV V NAATMMDK ...String: MKTHILLARV LACAALVLVT PGYSSEKIPV TGSGFVAKDD SLRTFFDAMA LQLKEPVIVS KMAARKKITG NFEFHDPNAL LEKLSLQLG LIWYFDGQAI YIYDASEMRN AVVSLRNVSL NEFNNFLKRS GLYNKNYPLR GDNRKGTFYV SGPPVYVDMV V NAATMMDK QNDGIELGRQ KIGVMRLNNT FVGDRTYNLR DQKMVIPGIA TAIERLLQGE EQPLGNIVSS EPPAMPAFSA NG EKGKAAN YAGGMSLQEA LKQNAAAGNI KIVAYPDTNS LLVKGTAEQV HFIEMLVKAL DVAKRHVELS LWIVDLNKSD LER LGTSWS GSITIGDKLG VSLNQSSIST LDGSRFIAAV NALEEKKQAT VVSRPVLLTQ ENVPAIFDNN RTFYTKLIGE RNVA LEHVT YGTMIRVLPR FSADGQIEMS LDIEDGNDKT PQSDTTTSVD ALPEVGRTLI STIARVPHGK SLLVGGYTRD ANTDT VQSI PFLGKLPLIG SLFRYSSKNK SNVVRVFMIE PKEIVDPLTP DASESVNNIL KQSGAWSGDD KLQKWVRVYL DRGQEA IK UniProtKB: SPI-1 type 3 secretion system secretin |
-Macromolecule #3: Protein PrgH
| Macromolecule | Name: Protein PrgH / type: protein_or_peptide / ID: 3 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 |
| Molecular weight | Theoretical: 44.509367 KDa |
| Sequence | String: METSKEKTIT SPGPYIVRLL NSSLNGCEFP LLTGRTLFVV GQSDALTASG QLPDIPADSF FIPLDHGGVN FEIQVDTDAT EIILHELKE GNSESRSVQL NTPIQVGELL ILIRPESEPW VPEQPEKLET SAKKNEPRFK NGIVAALAGF FILGIGTVGT L WILNSPQR ...String: METSKEKTIT SPGPYIVRLL NSSLNGCEFP LLTGRTLFVV GQSDALTASG QLPDIPADSF FIPLDHGGVN FEIQVDTDAT EIILHELKE GNSESRSVQL NTPIQVGELL ILIRPESEPW VPEQPEKLET SAKKNEPRFK NGIVAALAGF FILGIGTVGT L WILNSPQR QAAELDSLLG QEKERFQVLP GRDKMLYVAA QNERDTLWAR QVLARGDYDK NARVINENEE NKRISIWLDT YY PQLAYYR IHFDEPRKPV FWLSRQRNTM SKKELEVLSQ KLRALMPYAD SVNITLMDDV TAAGQAEAGL KQQALPYSRR NHK GGVTFV IQGALDDVEI LRARQFVDSY YRTWGGRYVQ FAIELKDDWL KGRSFQYGAE GYIKMSPGHW YFPSPL UniProtKB: Protein PrgH |
-Macromolecule #4: Surface presentation of antigens protein SpaP
| Macromolecule | Name: Surface presentation of antigens protein SpaP / type: protein_or_peptide / ID: 4 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 |
| Molecular weight | Theoretical: 25.249596 KDa |
| Sequence | String: MGNDISLIAL LAFSTLLPFI IASGTCFVKF SIVFVMVRNA LGLQQIPSNM TLNGVALLLS MFVMWPIMHD AYVYFEDEDV TFNDISSLS KHVDEGLDGY RDYLIKYSDR ELVQFFENAQ LKRQYGEETE TVKRDKDEIE KPSIFALLPA YALSEIKSAF K IGFYLYLP ...String: MGNDISLIAL LAFSTLLPFI IASGTCFVKF SIVFVMVRNA LGLQQIPSNM TLNGVALLLS MFVMWPIMHD AYVYFEDEDV TFNDISSLS KHVDEGLDGY RDYLIKYSDR ELVQFFENAQ LKRQYGEETE TVKRDKDEIE KPSIFALLPA YALSEIKSAF K IGFYLYLP FVVVDLVVSS VLLALGMMMM SPVTISTPIK LVLFVALDGW TLLSKGLILQ YMDIAT UniProtKB: Surface presentation of antigens protein SpaP |
-Macromolecule #5: Surface presentation of antigens protein SpaR
| Macromolecule | Name: Surface presentation of antigens protein SpaR / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 |
| Molecular weight | Theoretical: 28.499533 KDa |
| Sequence | String: MFYALYFEIH HLVASAALGF ARVAPIFFFL PFLNSGVLSG APRNAIIILV ALGVWPHALN EAPPFLSVAM IPLVLQEAAV GVMLGCLLS WPFWVMHALG CIIDNQRGAT LSSSIDPANG IDTSEMANFL NMFAAVVYLQ NGGLVTMVDV LNKSYQLCDP M NECTPSLP ...String: MFYALYFEIH HLVASAALGF ARVAPIFFFL PFLNSGVLSG APRNAIIILV ALGVWPHALN EAPPFLSVAM IPLVLQEAAV GVMLGCLLS WPFWVMHALG CIIDNQRGAT LSSSIDPANG IDTSEMANFL NMFAAVVYLQ NGGLVTMVDV LNKSYQLCDP M NECTPSLP PLLTFINQVA QNALVLASPV VLVLLLSEVF LGLLSRFAPQ MNAFAISLTV KSGIAVLIML LYFSPVLPDN VL RLSFQAT GLSSWFYERG ATHVLE UniProtKB: Surface presentation of antigens protein SpaR |
-Macromolecule #6: Surface presentation of antigens protein SpaQ
| Macromolecule | Name: Surface presentation of antigens protein SpaQ / type: protein_or_peptide / ID: 6 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 |
| Molecular weight | Theoretical: 9.363229 KDa |
| Sequence | String: MDDLVFAGNK ALYLVLILSG WPTIVATIIG LLVGLFQTVT QLQEQTLPFG IKLLGVCLCL FLLSGWYGEV LLSYGRQVIF LALAKG UniProtKB: Surface presentation of antigens protein SpaQ |
-Macromolecule #7: Protein PrgJ
| Macromolecule | Name: Protein PrgJ / type: protein_or_peptide / ID: 7 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 |
| Molecular weight | Theoretical: 10.934425 KDa |
| Sequence | String: MSIATIVPEN AVIGQAVNIR SMETDIVSLD DRLLQAFSGS AIATAVDKQT ITNRIEDPNL VTDPKELAIS QEMISDYNLY VSMVSTLTR KGVGAVETLL RS UniProtKB: Protein PrgJ |
-Macromolecule #8: Protein PrgI
| Macromolecule | Name: Protein PrgI / type: protein_or_peptide / ID: 8 / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria) Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) (bacteria)Strain: LT2 / SGSC1412 / ATCC 700720 |
| Molecular weight | Theoretical: 8.864868 KDa |
| Sequence | String: MATPWSGYLD DVSAKFDTGV DNLQTQVTEA LDKLAAKPSD PALLAAYQSK LSEYNLYRNA QSNTVKVFKD IDAAIIQNFR UniProtKB: SPI-1 type 3 secretion system needle filament protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 51.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 4.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 19141 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)