[English] 日本語
 Yorodumi
Yorodumi- EMDB-17189: Structure of the human neutral amino acid transporter ASCT2 in co... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human neutral amino acid transporter ASCT2 in complex with nanobody 469 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Neutral aminoacid transmembrane transporter activity / ASCT2 / Complex / Nanobody / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationglutamine secretion / L-glutamine import across plasma membrane / L-glutamine transmembrane transporter activity / glutamine transport / L-serine transmembrane transporter activity / ligand-gated channel activity / neutral amino acid transport / L-aspartate transmembrane transporter activity / L-aspartate import across plasma membrane / neutral L-amino acid transmembrane transporter activity ...glutamine secretion / L-glutamine import across plasma membrane / L-glutamine transmembrane transporter activity / glutamine transport / L-serine transmembrane transporter activity / ligand-gated channel activity / neutral amino acid transport / L-aspartate transmembrane transporter activity / L-aspartate import across plasma membrane / neutral L-amino acid transmembrane transporter activity / Amino acid transport across the plasma membrane / amino acid transmembrane transporter activity / symporter activity / antiporter activity / RHOJ GTPase cycle / protein homotrimerization / RHOQ GTPase cycle / amino acid transport / RHOH GTPase cycle / RAC3 GTPase cycle / transport across blood-brain barrier / RAC1 GTPase cycle / basal plasma membrane / erythrocyte differentiation / centriolar satellite / melanosome / signaling receptor activity / virus receptor activity / ciliary basal body / extracellular exosome / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
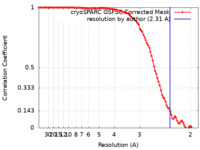
| Method | single particle reconstruction / cryo EM / Resolution: 2.31 Å | |||||||||
 Authors Authors | Canul-Tec J / Reyes N | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Receptor-recognition and antiviral mechanisms of retrovirus-derived human proteins. Authors: Shashank Khare / Miryam I Villalba / Juan C Canul-Tec / Arantza Balsebre Cajiao / Anand Kumar / Marija Backovic / Felix A Rey / Els Pardon / Jan Steyaert / Camilo Perez / Nicolas Reyes /     Abstract: Human syncytin-1 and suppressyn are cellular proteins of retroviral origin involved in cell-cell fusion events to establish the maternal-fetal interface in the placenta. In cell culture, they ...Human syncytin-1 and suppressyn are cellular proteins of retroviral origin involved in cell-cell fusion events to establish the maternal-fetal interface in the placenta. In cell culture, they restrict infections from members of the largest interference group of vertebrate retroviruses, and are regarded as host immunity factors expressed during development. At the core of the syncytin-1 and suppressyn functions are poorly understood mechanisms to recognize a common cellular receptor, the membrane transporter ASCT2. Here, we present cryo-electron microscopy structures of human ASCT2 in complexes with the receptor-binding domains of syncytin-1 and suppressyn. Despite their evolutionary divergence, the two placental proteins occupy similar positions in ASCT2, and are stabilized by the formation of a hybrid β-sheet or 'clamp' with the receptor. Structural predictions of the receptor-binding domains of extant retroviruses indicate overlapping binding interfaces and clamping sites with ASCT2, revealing a competition mechanism between the placental proteins and the retroviruses. Our work uncovers a common ASCT2 recognition mechanism by a large group of endogenous and disease-causing retroviruses, and provides high-resolution views on how placental human proteins exert morphological and immunological functions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17189.map.gz emd_17189.map.gz | 79 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17189-v30.xml emd-17189-v30.xml emd-17189.xml emd-17189.xml | 22.8 KB 22.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17189_fsc.xml emd_17189_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_17189.png emd_17189.png | 57.7 KB | ||
| Filedesc metadata |  emd-17189.cif.gz emd-17189.cif.gz | 7 KB | ||
| Others |  emd_17189_half_map_1.map.gz emd_17189_half_map_1.map.gz emd_17189_half_map_2.map.gz emd_17189_half_map_2.map.gz | 77.7 MB 77.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17189 http://ftp.pdbj.org/pub/emdb/structures/EMD-17189 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17189 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17189 | HTTPS FTP |
-Related structure data
| Related structure data |  8oudMC  8ouhC  8ouiC  8oujC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_17189.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17189.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.934 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_17189_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_17189_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of human ASCT2 with Nanobody 69
| Entire | Name: Complex of human ASCT2 with Nanobody 69 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of human ASCT2 with Nanobody 69
| Supramolecule | Name: Complex of human ASCT2 with Nanobody 69 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Molecular weight | Theoretical: 200 KDa |
-Supramolecule #2: Alanine Serine Cysteine Transporter 2
| Supramolecule | Name: Alanine Serine Cysteine Transporter 2 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Nanobody 69
| Supramolecule | Name: Nanobody 69 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Neutral amino acid transporter B(0)
| Macromolecule | Name: Neutral amino acid transporter B(0) / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.638902 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVADPPRDSK GLAAAEPTAN GGLALASIED QGAAAGGYCG SRDQVRRCLR ANLLVLLTVV AVVAGVALGL GVSGAGGALA LGPERLSAF VFPGELLLRL LRMIILPLVV CSLIGGAASL DPGALGRLGA WALLFFLVTT LLASALGVGL ALALQPGAAS A AINASVGA ...String: MVADPPRDSK GLAAAEPTAN GGLALASIED QGAAAGGYCG SRDQVRRCLR ANLLVLLTVV AVVAGVALGL GVSGAGGALA LGPERLSAF VFPGELLLRL LRMIILPLVV CSLIGGAASL DPGALGRLGA WALLFFLVTT LLASALGVGL ALALQPGAAS A AINASVGA AGSAENAPSK EVLDSFLDLA RNIFPSNLVS AAFRSYSTTY EERNITGTRV KVPVGQEVEG MNILGLVVFA IV FGVALRK LGPEGELLIR FFNSFNEATM VLVSWIMWYA PVGIMFLVAG KIVEMEDVGL LFARLGKYIL CCLLGHAIHG LLV LPLIYF LFTRKNPYRF LWGIVTPLAT AFGTSSSSAT LPLMMKCVEE NNGVAKHISR FILPIGATVN MDGAALFQCV AAVF IAQLS QQSLDFVKII TILVTATASS VGAAGIPAGG VLTLAIILEA VNLPVDHISL ILAVDWLVDR SCTVLNVEGD ALGAG LLQN YVDRTESRST EPELIQVKSE LPLDPLPVPT EEGNPLLKHY RGPAGDATVA SEKESVM UniProtKB: Neutral amino acid transporter B(0) |
-Macromolecule #2: Nanobody 469
| Macromolecule | Name: Nanobody 469 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.792409 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVESGGG LVQPGGSLRL SCAASGSIFR LDAMGWYRQA PGKQRELVAV IRSGGSTDYG DSVKGRFTIS RDNAKNTVYL QMNSLKPED TAVYYCNAVQ ILKTIYWGQG TQVTVSSHHH HHHEPEA |
-Macromolecule #3: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 3 / Number of copies: 9 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #4: ALANINE
| Macromolecule | Name: ALANINE / type: ligand / ID: 4 / Number of copies: 3 / Formula: ALA |
|---|---|
| Molecular weight | Theoretical: 89.093 Da |
| Chemical component information |  ChemComp-ALA: |
-Macromolecule #5: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 5 / Number of copies: 3 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 93 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8.1 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: 25 mM Hepes pH7.4, 100 mM NaCl, 5 mM L-Alanine, 0.05% DDS, 0.01% CHS. | ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 10 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.03 kPa | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 4344 / Average exposure time: 4.0 sec. / Average electron dose: 52.24 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 43-489 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 77.4 |
| Output model |  PDB-8oud: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)