[English] 日本語
 Yorodumi
Yorodumi- EMDB-16114: Vitamin B12 transporter BtuB1 with lipoprotein BtuG1 from B. theta -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vitamin B12 transporter BtuB1 with lipoprotein BtuG1 from B. theta | |||||||||
 Map data Map data | Sharpened with bfactor=-50 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Outer membrane / TonB-dependent transporter / vitamin B12 / lipoprotein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsiderophore transmembrane transport / siderophore uptake transmembrane transporter activity / cell outer membrane Similarity search - Function | |||||||||
| Biological species |  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) | |||||||||
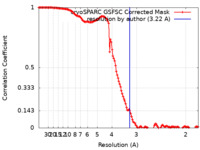
| Method | single particle reconstruction / cryo EM / Resolution: 3.22 Å | |||||||||
 Authors Authors | Silale A / Abellon-Ruiz J / van den Berg B | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: BtuB TonB-dependent transporters and BtuG surface lipoproteins form stable complexes for vitamin B uptake in gut Bacteroides. Authors: Javier Abellon-Ruiz / Kalyanashis Jana / Augustinas Silale / Andrew M Frey / Arnaud Baslé / Matthias Trost / Ulrich Kleinekathöfer / Bert van den Berg /   Abstract: Vitamin B (cobalamin) is required for most human gut microbes, many of which are dependent on scavenging to obtain this vitamin. Since bacterial densities in the gut are extremely high, competition ...Vitamin B (cobalamin) is required for most human gut microbes, many of which are dependent on scavenging to obtain this vitamin. Since bacterial densities in the gut are extremely high, competition for this keystone micronutrient is severe. Contrasting with Enterobacteria, members of the dominant genus Bacteroides often encode several BtuB vitamin B outer membrane transporters together with a conserved array of surface-exposed B-binding lipoproteins. Here we show that the BtuB transporters from Bacteroides thetaiotaomicron form stable, pedal bin-like complexes with surface-exposed BtuG lipoprotein lids, which bind B with high affinities. Closing of the BtuG lid following B capture causes destabilisation of the bound B by a conserved BtuB extracellular loop, causing translocation of the vitamin to BtuB and subsequent transport. We propose that TonB-dependent, lipoprotein-assisted small molecule uptake is a general feature of Bacteroides spp. that is important for the success of this genus in colonising the human gut. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16114.map.gz emd_16114.map.gz | 110.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16114-v30.xml emd-16114-v30.xml emd-16114.xml emd-16114.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16114_fsc.xml emd_16114_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_16114.png emd_16114.png | 65.1 KB | ||
| Masks |  emd_16114_msk_1.map emd_16114_msk_1.map | 216 MB |  Mask map Mask map | |
| Others |  emd_16114_additional_1.map.gz emd_16114_additional_1.map.gz emd_16114_half_map_1.map.gz emd_16114_half_map_1.map.gz emd_16114_half_map_2.map.gz emd_16114_half_map_2.map.gz | 108.3 MB 200.4 MB 200.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16114 http://ftp.pdbj.org/pub/emdb/structures/EMD-16114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16114 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16114 | HTTPS FTP |
-Validation report
| Summary document |  emd_16114_validation.pdf.gz emd_16114_validation.pdf.gz | 979.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_16114_full_validation.pdf.gz emd_16114_full_validation.pdf.gz | 979 KB | Display | |
| Data in XML |  emd_16114_validation.xml.gz emd_16114_validation.xml.gz | 21.4 KB | Display | |
| Data in CIF |  emd_16114_validation.cif.gz emd_16114_validation.cif.gz | 27.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16114 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16114 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16114 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-16114 | HTTPS FTP |
-Related structure data
| Related structure data |  8blwMC  8bmxC  8bmyC  8bmzC  8bn0C  8okvC  8p97C  8p98C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16114.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16114.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened with bfactor=-50 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
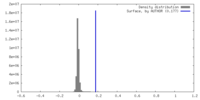
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16114_msk_1.map emd_16114_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
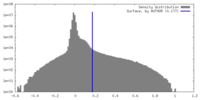
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_16114_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
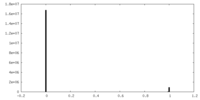
| Density Histograms |
-Half map: Half map A
| File | emd_16114_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
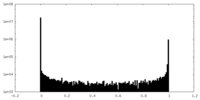
| Density Histograms |
-Half map: Half map B
| File | emd_16114_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of B. theta vitamin B12 transporter BtuB1 and lipoprotein...
| Entire | Name: Complex of B. theta vitamin B12 transporter BtuB1 and lipoprotein BtuG1 |
|---|---|
| Components |
|
-Supramolecule #1: Complex of B. theta vitamin B12 transporter BtuB1 and lipoprotein...
| Supramolecule | Name: Complex of B. theta vitamin B12 transporter BtuB1 and lipoprotein BtuG1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Molecular weight | Theoretical: 126 KDa |
-Macromolecule #1: Vitamin B12 transporter BtuB1
| Macromolecule | Name: Vitamin B12 transporter BtuB1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Molecular weight | Theoretical: 78.31275 KDa |
| Recombinant expression | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria) |
| Sequence | String: MRRNTFIKKM SVLLCCQALS LSLFAQQQKV DTAHIYSIPE IMVSDPYQTR EVRSASPLQV FNKEELKNLQ ALQVSDAVKH FAGVTVKDY GGIGGLKTVS IRSLGAQHTA VSYDGITVSD CQTGQVDIGR FSLNNVDRLS LNNGQSDNIF QPARFFASAG I LNIQTLTP ...String: MRRNTFIKKM SVLLCCQALS LSLFAQQQKV DTAHIYSIPE IMVSDPYQTR EVRSASPLQV FNKEELKNLQ ALQVSDAVKH FAGVTVKDY GGIGGLKTVS IRSLGAQHTA VSYDGITVSD CQTGQVDIGR FSLNNVDRLS LNNGQSDNIF QPARFFASAG I LNIQTLTP HFKEDKPTNI AAEFKTGSWG LVNPSLFLEQ QLNKKWSMTA NGEWMSSDGH YPFTLRYGND ADVQVSKEKR RN TDVENLR AEISAFANLS DKEQWRLKAY YYQSSRGLPN ATTLYYDFSR QNLRDKNTFI QSQYKKEFSR KWVFQTSAKW NWS YQNYQD PDALTSVGGT DNSYYQQEYY LSASALYRIW NNLSFSLSTD GSINTMNANL QNFVSPTRYS WLTAFAGKYV NEWV TLSAS ALATVINEKA KNGGNAGNHR KLSPNVSISL KPFHNEELRF RFFYKDIFRL PSFNDLYYDK AGNINLKPES ATQYN IGIT YSKAINNFIP YLSATVDAYH NKVTDKIVAT PTKNLFIWSM VNLGKVDIKG IDATASLSLQ PLDKLRINLS GNYTYQ RAL DVTNSNPNSP EGKVYKHQIA YTPRVSASGQ AGIETPWLNL SYSFLFSGKR YMLGQNISDN RLDSYSDHSI SAYRDFK IQ KVTASLNLEV LNLMNRNYEI VKNFPMPGRS VRVTIGVRYG GGHHHHHH UniProtKB: Vitamin B12 receptor, outer membrane |
-Macromolecule #2: YncE family protein
| Macromolecule | Name: YncE family protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Bacteroides thetaiotaomicron VPI-5482 (bacteria) Bacteroides thetaiotaomicron VPI-5482 (bacteria)Strain: ATCC 29148 / DSM 2079 / JCM 5827 / CCUG 10774 / NCTC 10582 / VPI-5482 / E50 |
| Molecular weight | Theoretical: 73.797047 KDa |
| Sequence | String: MQKGLLYNML LRLGKYFFLF PLFFIACDDL EDKPSIVPES NGDVFETGTA EMYILSEGLF NQNNSSLARY SFNRQRCTNN YFSANNQRG LGDTANDIAI YGNKIYVVVN VSSTVEVIDF PTGKSIRQIS MLRDNGSSRQ PRAIAFDKDK AYICSYDGTV A RIDTTSLE ...String: MQKGLLYNML LRLGKYFFLF PLFFIACDDL EDKPSIVPES NGDVFETGTA EMYILSEGLF NQNNSSLARY SFNRQRCTNN YFSANNQRG LGDTANDIAI YGNKIYVVVN VSSTVEVIDF PTGKSIRQIS MLRDNGSSRQ PRAIAFDKDK AYICSYDGTV A RIDTTSLE IEEIVTVGRN AEDICVQNGK LYVSNSGGLD YSGPGVDTTV SVIDITTFKE TKKIEVGPNP GKILPGLEEA VY VVTRGTD IEAGDYHLVK IDSRTDAVAI TYDEKVLSFA IDGPIAYLYT YDYQTKDSAI KVFDLNAGTV IRDNFITDGT AIQ TPFSIQ LNPFSGNIYI TEAYNYTVKG DVLCFNQQGQ LQYRLNDIGL NPNTVVFSDK ASQNEAGDTP EDPNAPSAFA NKVF EYIPA PGQFINTTTS AYEDGFSAGQ VLEHATEKLK KKSVISLGGF GGTITVGFHQ SIRNSKGEYD FRILGNASYN QNTGT GALG GSAEPGIVLV SKDENGNGLP DDEWYELAGS EYGKDTETRN YEITYYRPQP ANGDVRWTDN QGGEGFVYRN SYHQQD SYY PNWIEEDEIT FRGTRLKDNA INEGGTWVGY CYPWGYADNH PNRSEFSQFK IDWAVDQNGN HVELDKIDFV KIYTAVN QN VGWMGEISTE VMTVEDLHFE N UniProtKB: YncE family protein |
-Macromolecule #3: beta-D-galactopyranose
| Macromolecule | Name: beta-D-galactopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: GAL |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-GAL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 10 mM HEPES-NaOH, 100 mM NaCl, 0.05% DDM |
|---|---|
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number grids imaged: 1 / Number real images: 1924 / Average exposure time: 4.66 sec. / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 50 |
|---|---|
| Output model |  PDB-8blw: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)