[English] 日本語
 Yorodumi
Yorodumi- EMDB-16094: Cryo-EM structure of mouse heavy-chain apoferritin at 2.7 A plung... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mouse heavy-chain apoferritin at 2.7 A plunged 35ms after mixing with b-galactosidase | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | iron storage / ferritin / octahedral / METAL BINDING PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationIron uptake and transport / Golgi Associated Vesicle Biogenesis / negative regulation of ferroptosis / ferroxidase / autolysosome / ferroxidase activity / negative regulation of fibroblast proliferation / Neutrophil degranulation / endocytic vesicle lumen / ferric iron binding ...Iron uptake and transport / Golgi Associated Vesicle Biogenesis / negative regulation of ferroptosis / ferroxidase / autolysosome / ferroxidase activity / negative regulation of fibroblast proliferation / Neutrophil degranulation / endocytic vesicle lumen / ferric iron binding / autophagosome / iron ion transport / ferrous iron binding / intracellular iron ion homeostasis / immune response / iron ion binding / negative regulation of cell population proliferation / mitochondrion / extracellular region / identical protein binding / membrane / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | ||||||||||||
 Authors Authors | Torino S / Dhurandhar M / Efremov R | ||||||||||||
| Funding support | European Union,  Belgium, 3 items Belgium, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2023 Journal: Nat Methods / Year: 2023Title: Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting. Authors: Stefania Torino / Mugdha Dhurandhar / Annelore Stroobants / Raf Claessens / Rouslan G Efremov /  Abstract: Single-particle cryogenic electron microscopy (cryo-EM) allows reconstruction of high-resolution structures of proteins in different conformations. Protein function often involves transient ...Single-particle cryogenic electron microscopy (cryo-EM) allows reconstruction of high-resolution structures of proteins in different conformations. Protein function often involves transient functional conformations, which can be resolved using time-resolved cryo-EM (trEM). In trEM, reactions are arrested after a defined delay time by rapid vitrification of protein solution on the EM grid. Despite the increasing interest in trEM among the cryo-EM community, making trEM samples with a time resolution below 100 ms remains challenging. Here we report the design and the realization of a time-resolved cryo-plunger that combines a droplet-based microfluidic mixer with a laser-induced generator of microjets that allows rapid reaction initiation and plunge-freezing of cryo-EM grids. Using this approach, a time resolution of 5 ms was achieved and the protein density map was reconstructed to a resolution of 2.1 Å. trEM experiments on GroEL:GroES chaperonin complex resolved the kinetics of the complex formation and visualized putative short-lived conformations of GroEL-ATP complex. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16094.map.gz emd_16094.map.gz | 166.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16094-v30.xml emd-16094-v30.xml emd-16094.xml emd-16094.xml | 20.5 KB 20.5 KB | Display Display |  EMDB header EMDB header |
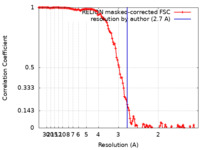
| FSC (resolution estimation) |  emd_16094_fsc.xml emd_16094_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16094.png emd_16094.png | 186.4 KB | ||
| Masks |  emd_16094_msk_1.map emd_16094_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16094.cif.gz emd-16094.cif.gz | 6.5 KB | ||
| Others |  emd_16094_half_map_1.map.gz emd_16094_half_map_1.map.gz emd_16094_half_map_2.map.gz emd_16094_half_map_2.map.gz | 61.3 MB 61.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16094 http://ftp.pdbj.org/pub/emdb/structures/EMD-16094 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16094 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16094 | HTTPS FTP |
-Related structure data
| Related structure data |  8bkaMC  8bk7C  8bk8C  8bk9C  8bkbC  8bkgC  8bkzC  8bl2C  8bl7C  8blcC  8bldC  8bleC  8blfC  8blyC  8bm0C  8bm1C  8bmdC  8bmoC  8bmtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16094.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16094.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.75 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16094_msk_1.map emd_16094_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16094_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16094_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Mouse heavy chain apoferritin from E.coli
| Entire | Name: Mouse heavy chain apoferritin from E.coli |
|---|---|
| Components |
|
-Supramolecule #1: Mouse heavy chain apoferritin from E.coli
| Supramolecule | Name: Mouse heavy chain apoferritin from E.coli / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Ferritin heavy chain
| Macromolecule | Name: Ferritin heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO / EC number: ferroxidase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.097631 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTTASPSQVR QNYHQDAEAA INRQINLELY ASYVYLSMSC YFDRDDVALK NFAKYFLHQS HEEREHAEKL MKLQNQRGGR IFLQDIKKP DRDDWESGLN AMECALHLEK SVNQSLLELH KLATDKNDPH LCDFIETYYL SEQVKSIKEL GDHVTNLRKM G APEAGMAE YLFDKHTLGH GDES UniProtKB: Ferritin heavy chain |
-Macromolecule #2: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 2 / Number of copies: 6 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 2271 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: contains Amaranth dye (acid red 27) 32 mM | ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 3 | ||||||||||||
| Vitrification | Cryogen name: ETHANE | ||||||||||||
| Details | Apoferritin in buffer of Amaranth dye (acid red 27 concentration 32 mM) |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 722 / Average exposure time: 2.796 sec. / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.55 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)