[English] 日本語
 Yorodumi
Yorodumi- EMDB-16157: Map of GroEL:GroES-(ADP) complex plunge frozen 200 ms after react... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Map of GroEL:GroES-(ADP) complex plunge frozen 200 ms after reaction initiation with ATP | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | GroEL / GroES / CHAPERONE | ||||||||||||
| Biological species |  | ||||||||||||
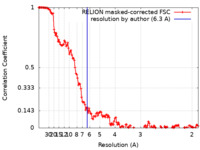
| Method | single particle reconstruction / cryo EM / Resolution: 6.3 Å | ||||||||||||
 Authors Authors | Dhurandhar M / Torino S / Efremov R | ||||||||||||
| Funding support | European Union,  Belgium, 3 items Belgium, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2023 Journal: Nat Methods / Year: 2023Title: Time-resolved cryo-EM using a combination of droplet microfluidics with on-demand jetting. Authors: Stefania Torino / Mugdha Dhurandhar / Annelore Stroobants / Raf Claessens / Rouslan G Efremov /  Abstract: Single-particle cryogenic electron microscopy (cryo-EM) allows reconstruction of high-resolution structures of proteins in different conformations. Protein function often involves transient ...Single-particle cryogenic electron microscopy (cryo-EM) allows reconstruction of high-resolution structures of proteins in different conformations. Protein function often involves transient functional conformations, which can be resolved using time-resolved cryo-EM (trEM). In trEM, reactions are arrested after a defined delay time by rapid vitrification of protein solution on the EM grid. Despite the increasing interest in trEM among the cryo-EM community, making trEM samples with a time resolution below 100 ms remains challenging. Here we report the design and the realization of a time-resolved cryo-plunger that combines a droplet-based microfluidic mixer with a laser-induced generator of microjets that allows rapid reaction initiation and plunge-freezing of cryo-EM grids. Using this approach, a time resolution of 5 ms was achieved and the protein density map was reconstructed to a resolution of 2.1 Å. trEM experiments on GroEL:GroES chaperonin complex resolved the kinetics of the complex formation and visualized putative short-lived conformations of GroEL-ATP complex. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16157.map.gz emd_16157.map.gz | 132.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16157-v30.xml emd-16157-v30.xml emd-16157.xml emd-16157.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16157_fsc.xml emd_16157_fsc.xml | 14 KB | Display |  FSC data file FSC data file |
| Images |  emd_16157.png emd_16157.png | 88.8 KB | ||
| Masks |  emd_16157_msk_1.map emd_16157_msk_1.map | 229.8 MB |  Mask map Mask map | |
| Others |  emd_16157_half_map_1.map.gz emd_16157_half_map_1.map.gz emd_16157_half_map_2.map.gz emd_16157_half_map_2.map.gz | 182.4 MB 182.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16157 http://ftp.pdbj.org/pub/emdb/structures/EMD-16157 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16157 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16157 | HTTPS FTP |
-Related structure data
| Related structure data |  8bk7C  8bk8C  8bk9C  8bkaC  8bkbC  8bkgC  8bkzC  8bl2C  8bl7C  8blcC  8bldC  8bleC  8blfC  8blyC  8bm0C  8bm1C  8bmdC  8bmoC  8bmtC C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_16157.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16157.map.gz / Format: CCP4 / Size: 229.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||||||||||||||||||
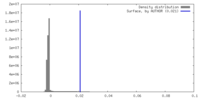
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16157_msk_1.map emd_16157_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
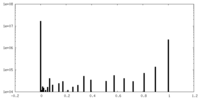
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_16157_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
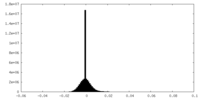
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_16157_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
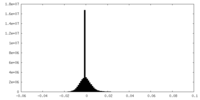
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Hetero 14mer assembled from 2 heptameric rings
| Entire | Name: Hetero 14mer assembled from 2 heptameric rings |
|---|---|
| Components |
|
-Supramolecule #1: Hetero 14mer assembled from 2 heptameric rings
| Supramolecule | Name: Hetero 14mer assembled from 2 heptameric rings / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 950 KDa |
-Macromolecule #1: GroEL
| Macromolecule | Name: GroEL / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MAAKDVKFGN DARVKMLRGV NVLADAVKVT LGPKGRNVVL DKSFGAPTIT KDGVSVAREI ELEDKFENMG AQMVKEVASK ANDAAGDGTT TATVLAQAII TEGLKAVAAG MNPMDLKRGI DKAVTAAVEE LKALSVPCSD SKAIAQVGTI SANSDETVGK LIAEAMDKVG ...String: MAAKDVKFGN DARVKMLRGV NVLADAVKVT LGPKGRNVVL DKSFGAPTIT KDGVSVAREI ELEDKFENMG AQMVKEVASK ANDAAGDGTT TATVLAQAII TEGLKAVAAG MNPMDLKRGI DKAVTAAVEE LKALSVPCSD SKAIAQVGTI SANSDETVGK LIAEAMDKVG KEGVITVEDG TGLQDELDVV EGMQFDRGYL SPYFINKPET GAVELESPFI LLADKKISNI REMLPVLEAV AKAGKPLLII AEDVEGEALA TLVVNTMRGI VKVAAVKAPG FGDRRKAMLQ DIATLTGGTV ISEEIGMELE KATLEDLGQA KRVVINKDTT TIIDGVGEEA AIQGRVAQIR QQIEEATSDY DREKLQERVA KLAGGVAVIK VGAATEVEMK EKKARVEDAL HATRAAVEEG VVAGGGVALI RVASKLADLR GQNEDQNVGI KVALRAMEAP LRQIVLNCGE EPSVVANTVK GGDGNYGYNA ATEEYGNMID MGILDPTKVT RSALQYAASV AGLMITTECM VTDLPKNDAA DLGAAGGMGG MGGMGGMM |
-Macromolecule #2: GroES
| Macromolecule | Name: GroES / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MANIRPLHDR VIVKRKEVET KSAGGIVLTG SAAAKSTRGE VLAVGNGRIL ENGEVKPLDV KVGDIVIFND GYGVKSEKID NEEVLIMSES DILAIVEA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS | ||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 4195 / Average electron dose: 63.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)