+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of yeast Elp456 subcomplex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | wobble uridine modification / TRANSLATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationelongator holoenzyme complex / protein urmylation / tRNA wobble uridine modification / tRNA modification / transcription elongation factor complex / peroxisome / regulation of translation / regulation of transcription by RNA polymerase II / nucleoplasm / identical protein binding ...elongator holoenzyme complex / protein urmylation / tRNA wobble uridine modification / tRNA modification / transcription elongation factor complex / peroxisome / regulation of translation / regulation of transcription by RNA polymerase II / nucleoplasm / identical protein binding / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
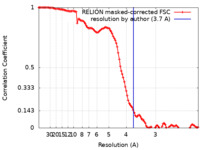
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Jaciuk M / Scherf D / Kaszuba K / Gaik M / Koscielniak A / Krutyholowa R / Rawski M / Indyka P / Biela A / Dobosz D ...Jaciuk M / Scherf D / Kaszuba K / Gaik M / Koscielniak A / Krutyholowa R / Rawski M / Indyka P / Biela A / Dobosz D / Lin T-Y / Abbassi N / Hammermeister A / Chramiec-Glabik A / Kosinski J / Schaffrath R / Glatt S | |||||||||
| Funding support |  Poland, European Union, 2 items Poland, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Cryo-EM structure of the fully assembled Elongator complex. Authors: Marcin Jaciuk / David Scherf / Karol Kaszuba / Monika Gaik / Alexander Rau / Anna Kościelniak / Rościsław Krutyhołowa / Michał Rawski / Paulina Indyka / Andrea Graziadei / Andrzej ...Authors: Marcin Jaciuk / David Scherf / Karol Kaszuba / Monika Gaik / Alexander Rau / Anna Kościelniak / Rościsław Krutyhołowa / Michał Rawski / Paulina Indyka / Andrea Graziadei / Andrzej Chramiec-Głąbik / Anna Biela / Dominika Dobosz / Ting-Yu Lin / Nour-El-Hana Abbassi / Alexander Hammermeister / Juri Rappsilber / Jan Kosinski / Raffael Schaffrath / Sebastian Glatt /    Abstract: Transfer RNA (tRNA) molecules are essential to decode messenger RNA codons during protein synthesis. All known tRNAs are heavily modified at multiple positions through post-transcriptional addition ...Transfer RNA (tRNA) molecules are essential to decode messenger RNA codons during protein synthesis. All known tRNAs are heavily modified at multiple positions through post-transcriptional addition of chemical groups. Modifications in the tRNA anticodons are directly influencing ribosome decoding and dynamics during translation elongation and are crucial for maintaining proteome integrity. In eukaryotes, wobble uridines are modified by Elongator, a large and highly conserved macromolecular complex. Elongator consists of two subcomplexes, namely Elp123 containing the enzymatically active Elp3 subunit and the associated Elp456 hetero-hexamer. The structure of the fully assembled complex and the function of the Elp456 subcomplex have remained elusive. Here, we show the cryo-electron microscopy structure of yeast Elongator at an overall resolution of 4.3 Å. We validate the obtained structure by complementary mutational analyses in vitro and in vivo. In addition, we determined various structures of the murine Elongator complex, including the fully assembled mouse Elongator complex at 5.9 Å resolution. Our results confirm the structural conservation of Elongator and its intermediates among eukaryotes. Furthermore, we complement our analyses with the biochemical characterization of the assembled human Elongator. Our results provide the molecular basis for the assembly of Elongator and its tRNA modification activity in eukaryotes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15635.map.gz emd_15635.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15635-v30.xml emd-15635-v30.xml emd-15635.xml emd-15635.xml | 23.7 KB 23.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15635_fsc.xml emd_15635_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_15635.png emd_15635.png | 135.3 KB | ||
| Filedesc metadata |  emd-15635.cif.gz emd-15635.cif.gz | 7.3 KB | ||
| Others |  emd_15635_half_map_1.map.gz emd_15635_half_map_1.map.gz emd_15635_half_map_2.map.gz emd_15635_half_map_2.map.gz | 104.7 MB 104.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15635 http://ftp.pdbj.org/pub/emdb/structures/EMD-15635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15635 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15635 | HTTPS FTP |
-Related structure data
| Related structure data |  8at6MC  8asvC  8aswC  8avgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15635.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15635.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_15635_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_15635_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Yeast Elp456 subcomplex
| Entire | Name: Yeast Elp456 subcomplex |
|---|---|
| Components |
|
-Supramolecule #1: Yeast Elp456 subcomplex
| Supramolecule | Name: Yeast Elp456 subcomplex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 233.8 KDa |
-Macromolecule #1: Elongator complex protein 4
| Macromolecule | Name: Elongator complex protein 4 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 51.232469 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSFRKRGEIL NDRGSGLRGP LLRGPPRTSS TPLRTGNRRA PGNVPLSDTT ARLKKLNIAD ESKTKMGLDS SHVGVRPSPA TSQPTTSTG SADLDSILGH MGLPLGNSVL VEEQSTTEFH SILGKLFAAQ GIVHNRISDS SADKTRNGDT HVIVLSLNQM F AKELPGIY ...String: MSFRKRGEIL NDRGSGLRGP LLRGPPRTSS TPLRTGNRRA PGNVPLSDTT ARLKKLNIAD ESKTKMGLDS SHVGVRPSPA TSQPTTSTG SADLDSILGH MGLPLGNSVL VEEQSTTEFH SILGKLFAAQ GIVHNRISDS SADKTRNGDT HVIVLSLNQM F AKELPGIY KGSRKQMKKN LISEEESKVT VQNLNETQRS TPSRYKDLKI AWKYKLADEK RLGSPDRDDI QQNSEYKDYN HQ FDITTRL MPAPIASELT FIAPTQPVST ILSQIEQTIK RNDKKLIRIV IPSLLHPAMY PPKMFESSEI IGLMHGVRSL VKK YYERVV LFASISIDII TPPLLVLLRN MFDSVINLEP FNQEMTEFLE RVYKSQPGKI QHGLVHILKL PVFTDRGEMR VLKS EWAFK NGRKKFEIEQ WGIPVDDAEG SAASEQSHSH SHSDEISHNI PAKKTKISLD Y UniProtKB: Elongator complex protein 4 |
-Macromolecule #2: Elongator complex protein 5
| Macromolecule | Name: Elongator complex protein 5 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 35.252496 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASSSHNPVI LLKRILSLTE SSPFILCLDS IAQTSYKLIQ EFVHQSKSKG NEYPIVYISF ETVNKPSYCT QFIDATQMDF VHLVKQIIS YLPAATATQA KKHMVIIDSL NYISTEYITR FLSEIASPHC TMVATYHKDI KDENRTVIPD WNNNYPDKLT L LQFMATTI ...String: MASSSHNPVI LLKRILSLTE SSPFILCLDS IAQTSYKLIQ EFVHQSKSKG NEYPIVYISF ETVNKPSYCT QFIDATQMDF VHLVKQIIS YLPAATATQA KKHMVIIDSL NYISTEYITR FLSEIASPHC TMVATYHKDI KDENRTVIPD WNNNYPDKLT L LQFMATTI VDIDVVLTGT LDTEEVSELL NEFRIPRGLN NDIFQLRLVN KRKSGRSLEY DFIVNSNTHE YELLSTTKQE EE SSSNGLE TPEMLQGLTT FNLGTSNKQK LAKDQVALPF LEAQSFGQGG AIVYEYEKDD DYDEEDPYED PF UniProtKB: Elongator complex protein 5 |
-Macromolecule #3: Elongator complex protein 6
| Macromolecule | Name: Elongator complex protein 6 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 30.602611 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSVQRQDLV LFSDQSVLPA HFFQDSNSHN LFFITHQSCT QPLWMINALV ETHVLGSPSS LNESSSSMLP SSTRSHAVLA SFIHEQNYF TNSLNKLKIP SNNYNVLDFL SDFIVNNIHN KPRDKILSDV LAKFSAAIQN NPTDTIVIIE QPELLLSLVS G LTCSELNN ...String: MGSVQRQDLV LFSDQSVLPA HFFQDSNSHN LFFITHQSCT QPLWMINALV ETHVLGSPSS LNESSSSMLP SSTRSHAVLA SFIHEQNYF TNSLNKLKIP SNNYNVLDFL SDFIVNNIHN KPRDKILSDV LAKFSAAIQN NPTDTIVIIE QPELLLSLVS G LTCSELNN KFITPLLRQC KVLIIVSNSD IFNIDEYDAS VHSSNLQNFY KSSFIKSMIN LNLNPLKTGF AKDVTGSLHV CR GGAPIAT SNTSLHVVEN EYLYLNEKES TKLFYR UniProtKB: Elongator complex protein 6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 15 s wait time, blot force 5, 5 s blot time. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number real images: 4716 / Average exposure time: 1.82 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)