[English] 日本語
 Yorodumi
Yorodumi- EMDB-15420: Structure of yeast oligosaccharylransferase complex with lipid-li... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of yeast oligosaccharylransferase complex with lipid-linked oligosaccharide and non-acceptor peptide bound | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | N-glycosylation OST complex / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationdolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / : / protein N-linked glycosylation / : / endoplasmic reticulum membrane / structural molecule activity / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Ramirez AS / de Capitani M / Pesciullesi G / Kowal J / Bloch JS / Irobalieva RN / Aebi M / Reymond JL / Locher KP | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Molecular basis for glycan recognition and reaction priming of eukaryotic oligosaccharyltransferase. Authors: Ana S Ramírez / Mario de Capitani / Giorgio Pesciullesi / Julia Kowal / Joël S Bloch / Rossitza N Irobalieva / Jean-Louis Reymond / Markus Aebi / Kaspar P Locher /  Abstract: Oligosaccharyltransferase (OST) is the central enzyme of N-linked protein glycosylation. It catalyzes the transfer of a pre-assembled glycan, GlcNAcManGlc, from a dolichyl-pyrophosphate donor to ...Oligosaccharyltransferase (OST) is the central enzyme of N-linked protein glycosylation. It catalyzes the transfer of a pre-assembled glycan, GlcNAcManGlc, from a dolichyl-pyrophosphate donor to acceptor sites in secretory proteins in the lumen of the endoplasmic reticulum. Precise recognition of the fully assembled glycan by OST is essential for the subsequent quality control steps of glycoprotein biosynthesis. However, the molecular basis of the OST-donor glycan interaction is unknown. Here we present cryo-EM structures of S. cerevisiae OST in distinct functional states. Our findings reveal that the terminal glucoses (Glc) of a chemo-enzymatically generated donor glycan analog bind to a pocket formed by the non-catalytic subunits WBP1 and OST2. We further find that binding either donor or acceptor substrate leads to distinct primed states of OST, where subsequent binding of the other substrate triggers conformational changes required for catalysis. This alternate priming allows OST to efficiently process closely spaced N-glycosylation sites. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15420.map.gz emd_15420.map.gz | 153.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15420-v30.xml emd-15420-v30.xml emd-15420.xml emd-15420.xml | 32.5 KB 32.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15420.png emd_15420.png | 61.3 KB | ||
| Filedesc metadata |  emd-15420.cif.gz emd-15420.cif.gz | 9.4 KB | ||
| Others |  emd_15420_half_map_1.map.gz emd_15420_half_map_1.map.gz emd_15420_half_map_2.map.gz emd_15420_half_map_2.map.gz | 151.5 MB 151.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15420 http://ftp.pdbj.org/pub/emdb/structures/EMD-15420 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15420 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15420 | HTTPS FTP |
-Related structure data
| Related structure data |  8agcMC  8agbC  8ageC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15420.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15420.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
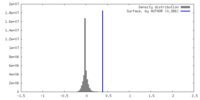
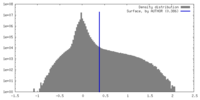
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_15420_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
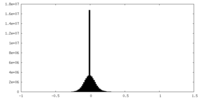
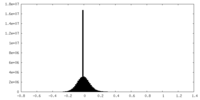
| Density Histograms |
-Half map: #1
| File | emd_15420_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
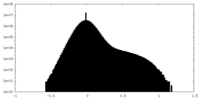
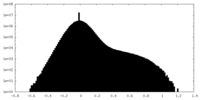
| Density Histograms |
- Sample components
Sample components
+Entire : Yeast OST complex with lipid-linked oligosaccharide and non-accep...
+Supramolecule #1: Yeast OST complex with lipid-linked oligosaccharide and non-accep...
+Macromolecule #1: Dolichyl-diphosphooligosaccharide--protein glycotransferase
+Macromolecule #2: OST4 isoform 1
+Macromolecule #3: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #4: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #5: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #6: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #7: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #8: OST3 isoform 1
+Macromolecule #9: PEPTIDE
+Macromolecule #13: phosphono [(3~{R},6~{E},10~{E})-3,7,11,15-tetramethylhexadeca-6,1...
+Macromolecule #14: 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #15: PHOSPHATIDYLETHANOLAMINE
+Macromolecule #16: MANGANESE (II) ION
+Macromolecule #17: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #18: 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)