[English] 日本語
 Yorodumi
Yorodumi- EMDB-15205: cryoEM structure of the catalytically inactive EndoS from S. pyog... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryoEM structure of the catalytically inactive EndoS from S. pyogenes in complex with the Fc region of immunoglobulin G1 | |||||||||
 Map data Map data | Main map cryosparc refine | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Endoglycosidase S / EndoS / endo-b-N-acetylglucosaminidase / Fc region / antibody / immunoglobulin G1 / Streptococcus pyogenes / N-glycans / HYDROLASE | |||||||||
| Function / homology | : / Endo-beta-N-acetylglucosaminidase F2, Ig-like domain / Glycosyl hydrolases family 18 (GH18) active site / Glycosyl hydrolases family 18 (GH18) active site signature. / hydrolase activity, hydrolyzing O-glycosyl compounds / Leucine-rich repeat domain superfamily / Glycoside hydrolase superfamily / carbohydrate metabolic process / Endo-beta-N-acetylglucosaminidase F2 Function and homology information Function and homology information | |||||||||
| Biological species |  Streptococcus pyogenes (bacteria) / Streptococcus pyogenes (bacteria) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
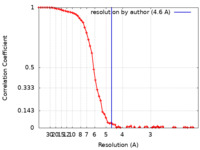
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Trastoy B / Cifuente JO / Du JJ / Sundberg EJ / Guerin ME | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Mechanism of antibody-specific deglycosylation and immune evasion by Streptococcal IgG-specific endoglycosidases. Authors: Beatriz Trastoy / Jonathan J Du / Javier O Cifuente / Lorena Rudolph / Mikel García-Alija / Erik H Klontz / Daniel Deredge / Nazneen Sultana / Chau G Huynh / Maria W Flowers / Chao Li / ...Authors: Beatriz Trastoy / Jonathan J Du / Javier O Cifuente / Lorena Rudolph / Mikel García-Alija / Erik H Klontz / Daniel Deredge / Nazneen Sultana / Chau G Huynh / Maria W Flowers / Chao Li / Diego E Sastre / Lai-Xi Wang / Francisco Corzana / Alvaro Mallagaray / Eric J Sundberg / Marcelo E Guerin /    Abstract: Bacterial pathogens have evolved intricate mechanisms to evade the human immune system, including the production of immunomodulatory enzymes. Streptococcus pyogenes serotypes secrete two multi- ...Bacterial pathogens have evolved intricate mechanisms to evade the human immune system, including the production of immunomodulatory enzymes. Streptococcus pyogenes serotypes secrete two multi-modular endo-β-N-acetylglucosaminidases, EndoS and EndoS2, that specifically deglycosylate the conserved N-glycan at Asn297 on IgG Fc, disabling antibody-mediated effector functions. Amongst thousands of known carbohydrate-active enzymes, EndoS and EndoS2 represent just a handful of enzymes that are specific to the protein portion of the glycoprotein substrate, not just the glycan component. Here, we present the cryoEM structure of EndoS in complex with the IgG1 Fc fragment. In combination with small-angle X-ray scattering, alanine scanning mutagenesis, hydrolytic activity measurements, enzyme kinetics, nuclear magnetic resonance and molecular dynamics analyses, we establish the mechanisms of recognition and specific deglycosylation of IgG antibodies by EndoS and EndoS2. Our results provide a rational basis from which to engineer novel enzymes with antibody and glycan selectivity for clinical and biotechnological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15205.map.gz emd_15205.map.gz | 10.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15205-v30.xml emd-15205-v30.xml emd-15205.xml emd-15205.xml | 24 KB 24 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15205_fsc.xml emd_15205_fsc.xml | 8.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_15205.png emd_15205.png | 92.3 KB | ||
| Masks |  emd_15205_msk_1.map emd_15205_msk_1.map | 22.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15205.cif.gz emd-15205.cif.gz | 8 KB | ||
| Others |  emd_15205_additional_1.map.gz emd_15205_additional_1.map.gz emd_15205_half_map_1.map.gz emd_15205_half_map_1.map.gz emd_15205_half_map_2.map.gz emd_15205_half_map_2.map.gz | 20.9 MB 20.6 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15205 http://ftp.pdbj.org/pub/emdb/structures/EMD-15205 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15205 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15205 | HTTPS FTP |
-Related structure data
| Related structure data |  8a64MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15205.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15205.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map cryosparc refine | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.052 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15205_msk_1.map emd_15205_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
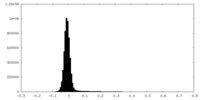
| Density Histograms |
-Additional map: Main map cryosparc refine
| File | emd_15205_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map cryosparc refine | ||||||||||||
| Projections & Slices |
| ||||||||||||
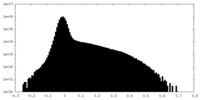
| Density Histograms |
-Half map: Half map cryosparc refine
| File | emd_15205_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map cryosparc refine | ||||||||||||
| Projections & Slices |
| ||||||||||||
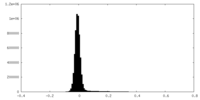
| Density Histograms |
-Half map: Half map cryosparc refine
| File | emd_15205_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map cryosparc refine | ||||||||||||
| Projections & Slices |
| ||||||||||||
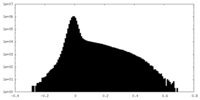
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of the EndoS from S. pyogenes and the Fc region of Immuno...
| Entire | Name: Complex of the EndoS from S. pyogenes and the Fc region of Immunoglobulin G |
|---|---|
| Components |
|
-Supramolecule #1: Complex of the EndoS from S. pyogenes and the Fc region of Immuno...
| Supramolecule | Name: Complex of the EndoS from S. pyogenes and the Fc region of Immunoglobulin G type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Endo-beta-N-acetylglucosaminidase F2
| Macromolecule | Name: Endo-beta-N-acetylglucosaminidase F2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 108.297039 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EEKTVQVQKG LPSIDSLHYL SENSKKEFKE ELSKAGQESQ KVKEILAKAQ QADKQAQELA KMKIPEKIPM KPLHGPLYGG YFRTWHDKT SDPTEKDKVN SMGELPKEVD LAFIFHDWTK DYSLFWKELA TKHVPKLNKQ GTRVIRTIPW RFLAGGDNSG I AEDTSKYP ...String: EEKTVQVQKG LPSIDSLHYL SENSKKEFKE ELSKAGQESQ KVKEILAKAQ QADKQAQELA KMKIPEKIPM KPLHGPLYGG YFRTWHDKT SDPTEKDKVN SMGELPKEVD LAFIFHDWTK DYSLFWKELA TKHVPKLNKQ GTRVIRTIPW RFLAGGDNSG I AEDTSKYP NTPEGNKALA KAIVDEYVYK YNLDGLDVDV AHDSIPKVDK KEDTAGVERS IQVFEEIGKL IGPKGVDKSR LF IMDSTYM ADKNPLIERG APYINLLLVQ VYGSQGEKGG WEPVSNRPEK TMEERWQGYS KYIRPEQYMI GFSFYEENAQ EGN LWYDIN SRKDEDKANG INTDITGTRA ERYARWQPKT GGVKGGIFSY AIDRDGVAHQ PKKYAKQKEF KDATDNIFHS DYSV SKALK TVMLKDKSYD LIDEKDFPDK ALREAVMAQV GTRKGDLERF NGTLRLDNPA IQSLEGLNKF KKLAQLDLIG LSRIT KLDR SVLPANMKPG KDTLETVLET YKKDNKEEPA TIPPVSLKVS GLTGLKELDL SGFDRETLAG LDAATLTSLE KVDISG NKL DLAPGTENRQ IFDTMLSTIS NHVGSNEQTV KFDKQKPTGH YPDTYGKTSL RLPVANEKVD LQSQLLFGTV TNQGTLI NS EADYKAYQNH KIAGRSFVDS NYHYNNFKVS YENYTVKVTD STLGTTTDKT LATDKEETYK VDFFSPADKT KAVHTAKV I VGDEKTMMVN LAEGATVIGG SADPVNARKV FDGQLGSETD NISLGWDSKQ SIIFKLKEDG LIKHWRFFND SARNPETTN KPIQEASLQI FNIKDYNLDN LLENPNKFDD EKYWITVDTY SAQGERATAF SNTLNNITSK YWRVVFDTKG DRYSSPVVPE LQILGYPLP NADTIMKTVT TAKELSQQKD KFSQKMLDEL KIKEMALETS LNSKIFDVTA INANAGVLKD CIEKRQLLKK L UniProtKB: Endo-beta-N-acetylglucosaminidase F2 |
-Macromolecule #2: Immunoglobulin gamma-1 heavy chain
| Macromolecule | Name: Immunoglobulin gamma-1 heavy chain / type: protein_or_peptide / ID: 2 Details: TCPPCPAPELLGGP SVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNS TYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDEL ...Details: TCPPCPAPELLGGP SVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNS TYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSRDEL TKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQ QGNVFSCSVMHEALHNHYTQKSLSLSPGK Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 49.386637 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLVQSGGG VVQPGRSLRL SCAASGFTFS RYTIHWVRQA PGKGLEWVAV MSYNGNNKHY ADSVNGRFTI SRNDSKNTLY LNMNSLRPE DTAVYYCARI RDTAMFFAHW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG ...String: QVQLVQSGGG VVQPGRSLRL SCAASGFTFS RYTIHWVRQA PGKGLEWVAV MSYNGNNKHY ADSVNGRFTI SRNDSKNTLY LNMNSLRPE DTAVYYCARI RDTAMFFAHW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG GTAALGCLVK DYFPEPVTVS W NSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKKVE PKSCDKTHTC PPCPAPELLG GP SVFLFPP KPKDTLMISR TPEVTCVVVD VSHEDPEVKF NWYVDGVEVH NAKTKPREEQ YNSTYRVVSV LTVLHQDWLN GKE YKCKVS NKALPAPIEK TISKAKGQPR EPQVYTLPPS RDELTKNQVS LTCLVKGFYP SDIAVEWESN GQPENNYKTT PPVL DSDGS FFLYSKLTVD KSRWQQGNVF SCSVMHEALH NHYTQKSLSL SPGK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: Standard PBS |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK II / Details: single blot. |
| Details | Complex stabilized by glutaraldehyde crosslinking prepared by ultracentrifugation in a fixation glycerol gradient (Grafix) and size exclusion chromatography purified at approximately 0.2 mg/mL |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 1 / Number real images: 5546 / Average electron dose: 59.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Details | Initial PDB models were rigid-body refined for individual chains. Later, the model was real-space refined removing side chains that couldn't be assigned. | ||||||||
| Refinement | Protocol: OTHER | ||||||||
| Output model |  PDB-8a64: |
 Movie
Movie Controller
Controller


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)