+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Ku heterodimer bound to DNA | |||||||||
 Map data Map data | Ku hetero-dimer bound to DNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NHEJ / Ku70 / Ku80 / DNA damage / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of lymphocyte differentiation / small-subunit processome assembly / Ku70:Ku80 complex / DNA-dependent protein kinase complex / negative regulation of t-circle formation / DNA end binding / DNA-dependent protein kinase-DNA ligase 4 complex / nonhomologous end joining complex / cellular response to X-ray / regulation of smooth muscle cell proliferation ...positive regulation of lymphocyte differentiation / small-subunit processome assembly / Ku70:Ku80 complex / DNA-dependent protein kinase complex / negative regulation of t-circle formation / DNA end binding / DNA-dependent protein kinase-DNA ligase 4 complex / nonhomologous end joining complex / cellular response to X-ray / regulation of smooth muscle cell proliferation / Cytosolic sensors of pathogen-associated DNA / DNA ligation / IRF3-mediated induction of type I IFN / nuclear telomere cap complex / double-strand break repair via classical nonhomologous end joining / positive regulation of catalytic activity / U3 snoRNA binding / recombinational repair / regulation of telomere maintenance / protein localization to chromosome, telomeric region / cellular response to fatty acid / positive regulation of neurogenesis / cellular hyperosmotic salinity response / hematopoietic stem cell proliferation / telomeric DNA binding / 2-LTR circle formation / : / site of DNA damage / Lyases; Carbon-oxygen lyases; Other carbon-oxygen lyases / 5'-deoxyribose-5-phosphate lyase activity / hematopoietic stem cell differentiation / positive regulation of protein kinase activity / ATP-dependent activity, acting on DNA / activation of innate immune response / enzyme activator activity / positive regulation of telomere maintenance via telomerase / DNA helicase activity / telomere maintenance / cyclin binding / neurogenesis / protein-DNA complex / cellular response to leukemia inhibitory factor / small-subunit processome / Nonhomologous End-Joining (NHEJ) / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / cellular response to gamma radiation / double-strand break repair via nonhomologous end joining / double-strand break repair / double-stranded DNA binding / scaffold protein binding / secretory granule lumen / DNA recombination / transcription regulator complex / ficolin-1-rich granule lumen / damaged DNA binding / chromosome, telomeric region / transcription cis-regulatory region binding / response to xenobiotic stimulus / ribonucleoprotein complex / innate immune response / negative regulation of DNA-templated transcription / DNA damage response / ubiquitin protein ligase binding / Neutrophil degranulation / protein-containing complex binding / nucleolus / positive regulation of DNA-templated transcription / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / protein-containing complex / DNA binding / RNA binding / extracellular region / nucleoplasm / ATP binding / membrane / nucleus / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.74 Å | |||||||||
 Authors Authors | Hardwick SW / Kefala-Stavridi A / Chirgadze DY / Blundell TL / Chaplin AK | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction. Authors: Antonia Kefala Stavridi / Amandine Gontier / Vincent Morin / Philippe Frit / Virginie Ropars / Nadia Barboule / Carine Racca / Sagun Jonchhe / Michael J Morten / Jessica Andreani / Alexey ...Authors: Antonia Kefala Stavridi / Amandine Gontier / Vincent Morin / Philippe Frit / Virginie Ropars / Nadia Barboule / Carine Racca / Sagun Jonchhe / Michael J Morten / Jessica Andreani / Alexey Rak / Pierre Legrand / Alexa Bourand-Plantefol / Steven W Hardwick / Dimitri Y Chirgadze / Paul Davey / Taiana Maia De Oliveira / Eli Rothenberg / Sebastien Britton / Patrick Calsou / Tom L Blundell / Paloma F Varela / Amanda K Chaplin / Jean-Baptiste Charbonnier /    Abstract: The classical Non-Homologous End Joining (c-NHEJ) pathway is the predominant process in mammals for repairing endogenous, accidental or programmed DNA Double-Strand Breaks. c-NHEJ is regulated by ...The classical Non-Homologous End Joining (c-NHEJ) pathway is the predominant process in mammals for repairing endogenous, accidental or programmed DNA Double-Strand Breaks. c-NHEJ is regulated by several accessory factors, post-translational modifications, endogenous chemical agents and metabolites. The metabolite inositol-hexaphosphate (IP6) stimulates c-NHEJ by interacting with the Ku70-Ku80 heterodimer (Ku). We report cryo-EM structures of apo- and DNA-bound Ku in complex with IP6, at 3.5 Å and 2.74 Å resolutions respectively, and an X-ray crystallography structure of a Ku in complex with DNA and IP6 at 3.7 Å. The Ku-IP6 interaction is mediated predominantly via salt bridges at the interface of the Ku70 and Ku80 subunits. This interaction is distant from the DNA, DNA-PKcs, APLF and PAXX binding sites and in close proximity to XLF binding site. Biophysical experiments show that IP6 binding increases the thermal stability of Ku by 2°C in a DNA-dependent manner, stabilizes Ku on DNA and enhances XLF affinity for Ku. In cells, selected mutagenesis of the IP6 binding pocket reduces both Ku accrual at damaged sites and XLF enrolment in the NHEJ complex, which translate into a lower end-joining efficiency. Thus, this study defines the molecular bases of the IP6 metabolite stimulatory effect on the c-NHEJ repair activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14986.map.gz emd_14986.map.gz | 194.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14986-v30.xml emd-14986-v30.xml emd-14986.xml emd-14986.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14986_fsc.xml emd_14986_fsc.xml | 17.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_14986.png emd_14986.png | 52.1 KB | ||
| Filedesc metadata |  emd-14986.cif.gz emd-14986.cif.gz | 7.1 KB | ||
| Others |  emd_14986_half_map_1.map.gz emd_14986_half_map_1.map.gz emd_14986_half_map_2.map.gz emd_14986_half_map_2.map.gz | 194.2 MB 194.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14986 http://ftp.pdbj.org/pub/emdb/structures/EMD-14986 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14986 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14986 | HTTPS FTP |
-Validation report
| Summary document |  emd_14986_validation.pdf.gz emd_14986_validation.pdf.gz | 939.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14986_full_validation.pdf.gz emd_14986_full_validation.pdf.gz | 939.1 KB | Display | |
| Data in XML |  emd_14986_validation.xml.gz emd_14986_validation.xml.gz | 20.6 KB | Display | |
| Data in CIF |  emd_14986_validation.cif.gz emd_14986_validation.cif.gz | 26.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14986 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14986 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14986 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14986 | HTTPS FTP |
-Related structure data
| Related structure data |  7zvtMC  7z6oC  7zt6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14986.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14986.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ku hetero-dimer bound to DNA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
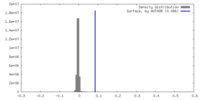
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_14986_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
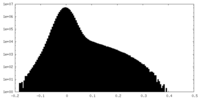
| Density Histograms |
-Half map: #1
| File | emd_14986_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
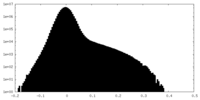
| Density Histograms |
- Sample components
Sample components
-Entire : Ku70/80 heterodimer bound to DNA
| Entire | Name: Ku70/80 heterodimer bound to DNA |
|---|---|
| Components |
|
-Supramolecule #1: Ku70/80 heterodimer bound to DNA
| Supramolecule | Name: Ku70/80 heterodimer bound to DNA / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3') type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.190741 KDa |
| Sequence | String: (DT)(DC)(DC)(DC)(DT)(DC)(DT)(DA)(DG)(DA) (DT)(DA)(DT)(DC) |
-Macromolecule #2: DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3') type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.96224 KDa |
| Sequence | String: (DC)(DG)(DA)(DT)(DA)(DT)(DC)(DT)(DA)(DG) (DA)(DG)(DG)(DG)(DA)(DT) |
-Macromolecule #3: X-ray repair cross-complementing protein 6
| Macromolecule | Name: X-ray repair cross-complementing protein 6 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 69.945039 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: MSGWESYYKT EGDEEAEEEQ EENLEASGDY KYSGRDSLIF LVDASKAMFE SQSEDELTPF DMSIQCIQSV YISKIISSDR DLLAVVFYG TEKDKNSVNF KNIYVLQELD NPGAKRILEL DQFKGQQGQK RFQDMMGHGS DYSLSEVLWV CANLFSDVQF K MSHKRIML ...String: MSGWESYYKT EGDEEAEEEQ EENLEASGDY KYSGRDSLIF LVDASKAMFE SQSEDELTPF DMSIQCIQSV YISKIISSDR DLLAVVFYG TEKDKNSVNF KNIYVLQELD NPGAKRILEL DQFKGQQGQK RFQDMMGHGS DYSLSEVLWV CANLFSDVQF K MSHKRIML FTNEDNPHGN DSAKASRART KAGDLRDTGI FLDLMHLKKP GGFDISLFYR DIISIAEDED LRVHFEESSK LE DLLRKVR AKETRKRALS RLKLKLNKDI VISVGIYNLV QKALKPPPIK LYRETNEPVK TKTRTFNTST GGLLLPSDTK RSQ IYGSRQ IILEKEETEE LKRFDDPGLM LMGFKPLVLL KKHHYLRPSL FVYPEESLVI GSSTLFSALL IKCLEKEVAA LCRY TPRRN IPPYFVALVP QEEELDDQKI QVTPPGFQLV FLPFADDKRK MPFTEKIMAT PEQVGKMKAI VEKLRFTYRS DSFEN PVLQ QHFRNLEALA LDLMEPEQAV DLTLPKVEAM NKRLGSLVDE FKELVYPPDY NPEGKVTKRK HDNEGSGSKR PKVEYS EEE LKTHISKGTL GKFTVPMLKE ACRAYGLKSG LKKQELLEAL TKHFQD UniProtKB: X-ray repair cross-complementing protein 6 |
-Macromolecule #4: X-ray repair cross-complementing protein 5
| Macromolecule | Name: X-ray repair cross-complementing protein 5 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 82.812438 KDa |
| Recombinant expression | Organism: Insect cell expression vector pTIE1 (others) |
| Sequence | String: MVRSGNKAAV VLCMDVGFTM SNSIPGIESP FEQAKKVITM FVQRQVFAEN KDEIALVLFG TDGTDNPLSG GDQYQNITVH RHLMLPDFD LLEDIESKIQ PGSQQADFLD ALIVSMDVIQ HETIGKKFEK RHIEIFTDLS SRFSKSQLDI IIHSLKKCDI S LQFFLPFS ...String: MVRSGNKAAV VLCMDVGFTM SNSIPGIESP FEQAKKVITM FVQRQVFAEN KDEIALVLFG TDGTDNPLSG GDQYQNITVH RHLMLPDFD LLEDIESKIQ PGSQQADFLD ALIVSMDVIQ HETIGKKFEK RHIEIFTDLS SRFSKSQLDI IIHSLKKCDI S LQFFLPFS LGKEDGSGDR GDGPFRLGGH GPSFPLKGIT EQQKEGLEIV KMVMISLEGE DGLDEIYSFS ESLRKLCVFK KI ERHSIHW PCRLTIGSNL SIRIAAYKSI LQERVKKTWT VVDAKTLKKE DIQKETVYCL NDDDETEVLK EDIIQGFRYG SDI VPFSKV DEEQMKYKSE GKCFSVLGFC KSSQVQRRFF MGNQVLKVFA ARDDEAAAVA LSSLIHALDD LDMVAIVRYA YDKR ANPQV GVAFPHIKHN YECLVYVQLP FMEDLRQYMF SSLKNSKKYA PTEAQLNAVD ALIDSMSLAK KDEKTDTLED LFPTT KIPN PRFQRLFQCL LHRALHPREP LPPIQQHIWN MLNPPAEVTT KSQIPLSKIK TLFPLIEAKK KDQVTAQEIF QDNHED GPT AKKLKTEQGG AHFSVSSLAE GSVTSVGSVN PAENFRVLVK QKKASFEEAS NQLINHIEQF LDTNETPYFM KSIDCIR AF REEAIKFSEE QRFNNFLKAL QEKVEIKQLN HFWEIVVQDG ITLITKEEAS GSSVTAEEAK KFLAPKDKPS GDTAAVFE E GGDVDDLLDM I UniProtKB: X-ray repair cross-complementing protein 5 |
-Macromolecule #5: INOSITOL HEXAKISPHOSPHATE
| Macromolecule | Name: INOSITOL HEXAKISPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: IHP |
|---|---|
| Molecular weight | Theoretical: 660.035 Da |
| Chemical component information |  ChemComp-IHP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6924 / Average exposure time: 1.25 sec. / Average electron dose: 44.39 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)