[English] 日本語
 Yorodumi
Yorodumi- EMDB-14849: Nup93 in complex with 5-Nup93 inhibitory NB and 15-Nup93 tracking NB -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nup93 in complex with 5-Nup93 inhibitory NB and 15-Nup93 tracking NB | |||||||||
 Map data Map data | Nup93 in complex with inhibitory Nanobody | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NUP93 / Nuclearporin / inhibitory NB / tracking NB / NUCLEAR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnuclear pore complex assembly / structural constituent of nuclear pore / poly(A)+ mRNA export from nucleus / nuclear pore / nuclear periphery / protein import into nucleus / nuclear membrane / cytosol Similarity search - Function | |||||||||
| Biological species | Xenopus laevis, Vicugna pacos /  | |||||||||
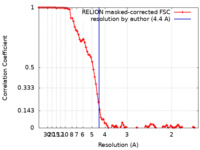
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Fu Z / Guttler T / Colom MS | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2024 Journal: EMBO J / Year: 2024Title: A checkpoint function for Nup98 in nuclear pore formation suggested by novel inhibitory nanobodies. Authors: Mireia Solà Colom / Zhenglin Fu / Philip Gunkel / Thomas Güttler / Sergei Trakhanov / Vasundara Srinivasan / Kathrin Gregor / Tino Pleiner / Dirk Görlich /   Abstract: Nuclear pore complex (NPC) biogenesis is a still enigmatic example of protein self-assembly. We now introduce several cross-reacting anti-Nup nanobodies for imaging intact nuclear pore complexes from ...Nuclear pore complex (NPC) biogenesis is a still enigmatic example of protein self-assembly. We now introduce several cross-reacting anti-Nup nanobodies for imaging intact nuclear pore complexes from frog to human. We also report a simplified assay that directly tracks postmitotic NPC assembly with added fluorophore-labeled anti-Nup nanobodies. During interphase, NPCs are inserted into a pre-existing nuclear envelope. Monitoring this process is challenging because newly assembled NPCs are indistinguishable from pre-existing ones. We overcame this problem by inserting Xenopus-derived NPCs into human nuclear envelopes and using frog-specific anti-Nup nanobodies for detection. We further asked whether anti-Nup nanobodies could serve as NPC assembly inhibitors. Using a selection strategy against conserved epitopes, we obtained anti-Nup93, Nup98, and Nup155 nanobodies that block Nup-Nup interfaces and arrest NPC assembly. We solved structures of nanobody-target complexes and identified roles for the Nup93 α-solenoid domain in recruiting Nup358 and the Nup214·88·62 complex, as well as for Nup155 and the Nup98 autoproteolytic domain in NPC scaffold assembly. The latter suggests a checkpoint linking pore formation to the assembly of the Nup98-dominated permeability barrier. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14849.map.gz emd_14849.map.gz | 4.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14849-v30.xml emd-14849-v30.xml emd-14849.xml emd-14849.xml | 20.8 KB 20.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14849_fsc.xml emd_14849_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14849.png emd_14849.png | 58.3 KB | ||
| Filedesc metadata |  emd-14849.cif.gz emd-14849.cif.gz | 6.7 KB | ||
| Others |  emd_14849_half_map_1.map.gz emd_14849_half_map_1.map.gz emd_14849_half_map_2.map.gz emd_14849_half_map_2.map.gz | 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14849 http://ftp.pdbj.org/pub/emdb/structures/EMD-14849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14849 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14849 | HTTPS FTP |
-Related structure data
| Related structure data |  7zoxMC  7nowC  7nqaC  8cdsC  8cdtC  8ozbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14849.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14849.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nup93 in complex with inhibitory Nanobody | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.834 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map of Nup93 in complex with inhibitory Nanobody
| File | emd_14849_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of Nup93 in complex with inhibitory Nanobody | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map of Nup93 in complex with inhibitory Nanobody
| File | emd_14849_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map of Nup93 in complex with inhibitory Nanobody | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nup93 in complex with 5-Nup93 inhibitory NB and 15-Nup93 tracking NB
| Entire | Name: Nup93 in complex with 5-Nup93 inhibitory NB and 15-Nup93 tracking NB |
|---|---|
| Components |
|
-Supramolecule #1: Nup93 in complex with 5-Nup93 inhibitory NB and 15-Nup93 tracking NB
| Supramolecule | Name: Nup93 in complex with 5-Nup93 inhibitory NB and 15-Nup93 tracking NB type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism: Xenopus laevis, Vicugna pacos |
| Molecular weight | Theoretical: 100 KDa |
-Macromolecule #1: Nuclear pore complex protein Nup93
| Macromolecule | Name: Nuclear pore complex protein Nup93 / type: protein_or_peptide / ID: 1 / Details: Xenopus laevis Nup93(residues 168-820) / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 74.782289 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SESGAPGRSS LDNVEMAYAR QMYMYNEKVV SGHLQPSLVD LCTEAAERLD DKNVSDLWVM VKQMTDVPLI PASDTLKSRC SGQMQMAFV RQALNYLEQS YKNYTLISVF ANLQQAQLGG VPGTYNLVRS FLNIRLPTPI PGLQDGEIEG YPVWALIYYC M RCGDLMAA ...String: SESGAPGRSS LDNVEMAYAR QMYMYNEKVV SGHLQPSLVD LCTEAAERLD DKNVSDLWVM VKQMTDVPLI PASDTLKSRC SGQMQMAFV RQALNYLEQS YKNYTLISVF ANLQQAQLGG VPGTYNLVRS FLNIRLPTPI PGLQDGEIEG YPVWALIYYC M RCGDLMAA QQVVNRAQHQ LGDFKNCFQE YIHNKDRRLS PTTENKLRLH YRRAVRASTD PYKRAVYCII GRCDVSDNHS EV ADKTEDY LWLKLSQVCF EDEANSSPQD RLTLPQFQKQ LFEDYGESHF AVNQQPYLYF QVLFLTAQFE AAIAFLFRLE RTR CHAVHV ALALFELKLL LKSTGQSAQL LSQEPGEPQG VRRLNFIRLL MLYTRKFEPT DPREALQYFY FLRNEKDNQG ESMF LRCVS ELVIESREFD MLLGKLEKDG SRKPGAIDKF TRDTKTIINK VASVAENKGL FEEAAKLYDL AKNPDKVLEL TNKLL SPVV SQISAPQSNR ERLKNMALAI AERYKSQGVS AEKSINSTFY LLLDLITFFD EYHAGHIDLS FDVIERLKLV PLSQDS VEE RVAAFRNFSD EIRHNLSEIL LATMNILFTQ YKRLKGSGPT TLGRPQRVQE DKDSVLRSQA RALITFAGMI PYRMSGD TN ARLVQMEVLM N UniProtKB: Nuclear pore complex protein Nup93 |
-Macromolecule #2: xhNup93-Nb4i
| Macromolecule | Name: xhNup93-Nb4i / type: protein_or_peptide / ID: 2 / Details: 5-Nup93 inhibitory NB / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.811384 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GQVQLVESGG GVVQTGDSLM LSCTVSGHTI DNWAMGWFRQ TPRKQREFVA AIDKRGTGAI YGNSVKGRFT VSRDNAKNMV YLRMNSLKP EDTAVYFCAV DQLNAGLGDV SYDYDYWGQG TQVTVSS |
-Macromolecule #3: xNup93-Nb2t
| Macromolecule | Name: xNup93-Nb2t / type: protein_or_peptide / ID: 3 / Details: 15-Nup93 tracking NB / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.921641 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GDVQLVESGG GLVQPGGSLR LSCVASGFTF SRVGMGWVRQ APGKGLEWVS DINASGGTGY ADSVKGRFAI SRDNAKNTLY LQMNRLKPE DTAVYYCAKM MDTAMIEAGI IKPAGQGTQV TVSS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.04 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Sample volume: 2.0 microliters blotting time: 5 s blot force setting: 6. | ||||||||||||
| Details | The complex was further purified by size exclusion chromatography using a HiLoad 26/60 Superdex 200 column. The purified complex was applied to a glow-discharged grid after being diluted to 1 mg/ml. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 12320 / Average exposure time: 2.0 sec. / Average electron dose: 54.0 e/Å2 / Details: Counting mode 5 images per hole( beam-image shift) |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)