+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13871 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | ||||||||||||
 Map data Map data | Single particle Cryo-EM map of Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | SARS-CoV2 / Spike / glycoprotein / antibody / fab / B.1.135 / beta variant / Complex / neutralising / convalescent sera / Viral Protein/Immune System / Viral Protein-Immune System complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | ||||||||||||
 Authors Authors | Duyvesteyn HME / Ren J / Stuart DI | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell Host Microbe / Year: 2022 Journal: Cell Host Microbe / Year: 2022Title: The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants. Authors: Chang Liu / Daming Zhou / Rungtiwa Nutalai / Helen M E Duyvesteyn / Aekkachai Tuekprakhon / Helen M Ginn / Wanwisa Dejnirattisai / Piyada Supasa / Alexander J Mentzer / Beibei Wang / James ...Authors: Chang Liu / Daming Zhou / Rungtiwa Nutalai / Helen M E Duyvesteyn / Aekkachai Tuekprakhon / Helen M Ginn / Wanwisa Dejnirattisai / Piyada Supasa / Alexander J Mentzer / Beibei Wang / James Brett Case / Yuguang Zhao / Donal T Skelly / Rita E Chen / Sile Ann Johnson / Thomas G Ritter / Chris Mason / Tariq Malik / Nigel Temperton / Neil G Paterson / Mark A Williams / David R Hall / Daniel K Clare / Andrew Howe / Philip J R Goulder / Elizabeth E Fry / Michael S Diamond / Juthathip Mongkolsapaya / Jingshan Ren / David I Stuart / Gavin R Screaton /    Abstract: Alpha-B.1.1.7, Beta-B.1.351, Gamma-P.1, and Delta-B.1.617.2 variants of SARS-CoV-2 express multiple mutations in the spike protein (S). These may alter the antigenic structure of S, causing escape ...Alpha-B.1.1.7, Beta-B.1.351, Gamma-P.1, and Delta-B.1.617.2 variants of SARS-CoV-2 express multiple mutations in the spike protein (S). These may alter the antigenic structure of S, causing escape from natural or vaccine-induced immunity. Beta is particularly difficult to neutralize using serum induced by early pandemic SARS-CoV-2 strains and is most antigenically separated from Delta. To understand this, we generated 674 mAbs from Beta-infected individuals and performed a detailed structure-function analysis of the 27 most potent mAbs: one binding the spike N-terminal domain (NTD), the rest the receptor-binding domain (RBD). Two of these RBD-binding mAbs recognize a neutralizing epitope conserved between SARS-CoV-1 and -2, while 18 target mutated residues in Beta: K417N, E484K, and N501Y. There is a major response to N501Y, including a public IgVH4-39 sequence, with E484K and K417N also targeted. Recognition of these key residues underscores why serum from Beta cases poorly neutralizes early pandemic and Delta viruses. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13871.map.gz emd_13871.map.gz | 313.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13871-v30.xml emd-13871-v30.xml emd-13871.xml emd-13871.xml | 17.6 KB 17.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13871.png emd_13871.png | 21.4 KB | ||
| Filedesc metadata |  emd-13871.cif.gz emd-13871.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13871 http://ftp.pdbj.org/pub/emdb/structures/EMD-13871 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13871 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13871 | HTTPS FTP |
-Related structure data
| Related structure data |  7q9jMC  7pryC  7przC  7ps0C  7ps1C  7ps2C  7ps3C  7ps4C  7ps5C  7ps6C  7ps7C  7q0gC  7q0hC  7q0iC  7q6eC  7q9fC  7q9gC  7q9iC  7q9kC  7q9mC  7q9pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13871.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13871.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single particle Cryo-EM map of Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
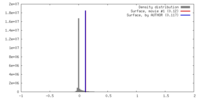
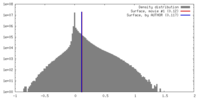
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
| Entire | Name: Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein |
|---|---|
| Components |
|
-Supramolecule #1: Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein
| Supramolecule | Name: Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 475 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 141.993984 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNFTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFA NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF ...String: MFVFLVLLPL VSSQCVNFTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFA NPVLPFNDG VYFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV Y SSANNCTF EYVSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRGLPQ GFSALEPLVD LPIGINITRF QT LHISYLT PGDSSSGWTA GAAAYYVGYL QPRTFLLKYN ENGTITDAVD CALDPLSETK CTLKSFTVEK GIYQTSNFRV QPT ESIVRF PNITNLCPFG EVFNATRFAS VYAWNRKRIS NCVADYSVLY NSASFSTFKC YGVSPTKLND LCFTNVYADS FVIR GDEVR QIAPGQTGNI ADYNYKLPDD FTGCVIAWNS NNLDSKVGGN YNYLYRLFRK SNLKPFERDI STEIYQAGST PCNGV KGFN CYFPLQSYGF QPTYGVGYQP YRVVVLSFEL LHAPATVCGP KKSTNLVKNK CVNFNFNGLT GTGVLTESNK KFLPFQ QFG RDIADTTDAV RDPQTLEILD ITPCSFGGVS VITPGTNTSN QVAVLYQGVN CTEVPVAIHA DQLTPTWRVY STGSNVF QT RAGCLIGAEH VNNSYECDIP IGAGICASYQ TQTNSPGSAS SVASQSIIAY TMSLGVENSV AYSNNSIAIP TNFTISVT T EILPVSMTKT SVDCTMYICG DSTECSNLLL QYGSFCTQLN RALTGIAVEQ DKNTQEVFAQ VKQIYKTPPI KDFGGFNFS QILPDPSKPS KRSFIEDLLF NKVTLADAGF IKQYGDCLGD IAARDLICAQ KFNGLTVLPP LLTDEMIAQY TSALLAGTIT SGWTFGAGA ALQIPFAMQM AYRFNGIGVT QNVLYENQKL IANQFNSAIG KIQDSLSSTA SALGKLQDVV NQNAQALNTL V KQLSSNFG AISSVLNDIL SRLDPPEAEV QIDRLITGRL QSLQTYVTQQ LIRAAEIRAS ANLAATKMSE CVLGQSKRVD FC GKGYHLM SFPQSAPHGV VFLHVTYVPA QEKNFTTAPA ICHDGKAHFP REGVFVSNGT HWFVTQRNFY EPQIITTDNT FVS GNCDVV IGIVNNTVYD PLQPELDSFK EELDKYFKNH TSPDVDLGDI SGINASVVNI QKEIDRLNEV AKNLNESLID LQEL GKYEQ GSGYIPEAPR DGQAYVRKDG EWVLLSTFLG RSLEVLFQGP GHHHHHHHHG SAWSHPQFEK GGGSGGGSGG SAWSH PQFE K UniProtKB: Spike glycoprotein |
-Macromolecule #2: Beta-26 light chain
| Macromolecule | Name: Beta-26 light chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 22.706889 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SYELTQPPSV SVAPGQTARI TCGGNNIGSK SVHWFQQKPG QAPVLVVYDD SDRPSGIPER FSGSNSGNTA SLTISRVEAG DEADYYCQV WDSASDSGVF GTGTKLTVPG QPKANPTVTL FPPSSEELQA NKATLVCLIS DFYPGAVTVA WKADSSPVKA G VETTTPSK ...String: SYELTQPPSV SVAPGQTARI TCGGNNIGSK SVHWFQQKPG QAPVLVVYDD SDRPSGIPER FSGSNSGNTA SLTISRVEAG DEADYYCQV WDSASDSGVF GTGTKLTVPG QPKANPTVTL FPPSSEELQA NKATLVCLIS DFYPGAVTVA WKADSSPVKA G VETTTPSK QSNNKYAASS YLSLTPEQWK SHRSYSCQVT HEGSTVEKTV APTECS |
-Macromolecule #3: Beta-26 heavy chain
| Macromolecule | Name: Beta-26 heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.104998 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QVQLQQSGPG LVKPSETLSL TCTVSGASIS NYYWSWIRQP PGKGLEWVGY IYYTGSTNHN PSLKSRVTIS LDTSKNQFSL RLSSVTAAD TAVYYCARAY CSGGSCFDTF DIWGQGTMVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL ...String: QVQLQQSGPG LVKPSETLSL TCTVSGASIS NYYWSWIRQP PGKGLEWVGY IYYTGSTNHN PSLKSRVTIS LDTSKNQFSL RLSSVTAAD TAVYYCARAY CSGGSCFDTF DIWGQGTMVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV T VSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSCDK |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 31 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: C-flat-2/1 / Material: COPPER/RHODIUM / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average electron dose: 40.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7q9j: |
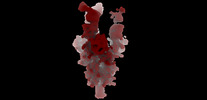
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)