+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13045 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Brr2 in complex with Fbp21 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mRNA Splicing / Spliceosomal Assembly / Splicing Regulation / Brr2 Helicase / SPLICING | |||||||||
| Function / homology |  Function and homology information Function and homology informationcis assembly of pre-catalytic spliceosome / spliceosome conformational change to release U4 (or U4atac) and U1 (or U11) / U2-type catalytic step 1 spliceosome / U2-type precatalytic spliceosome / proline-rich region binding / mRNA cis splicing, via spliceosome / precatalytic spliceosome / mRNA Splicing - Minor Pathway / U5 snRNP / U4/U6 x U5 tri-snRNP complex ...cis assembly of pre-catalytic spliceosome / spliceosome conformational change to release U4 (or U4atac) and U1 (or U11) / U2-type catalytic step 1 spliceosome / U2-type precatalytic spliceosome / proline-rich region binding / mRNA cis splicing, via spliceosome / precatalytic spliceosome / mRNA Splicing - Minor Pathway / U5 snRNP / U4/U6 x U5 tri-snRNP complex / catalytic step 2 spliceosome / mRNA Splicing - Major Pathway / RNA splicing / helicase activity / spliceosomal complex / mRNA splicing, via spliceosome / osteoblast differentiation / RNA helicase activity / RNA helicase / nuclear speck / ATP hydrolysis activity / RNA binding / zinc ion binding / nucleoplasm / ATP binding / identical protein binding / membrane / nucleus Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
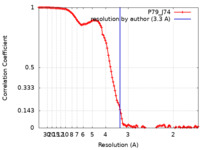
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Bergfort A / Hilal T | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling. Authors: Alexandra Bergfort / Tarek Hilal / Benno Kuropka / İbrahim Avşar Ilik / Gert Weber / Tuğçe Aktaş / Christian Freund / Markus C Wahl /  Abstract: Biogenesis of spliceosomal small nuclear ribonucleoproteins (snRNPs) and their recycling after splicing require numerous assembly/recycling factors whose modes of action are often poorly understood. ...Biogenesis of spliceosomal small nuclear ribonucleoproteins (snRNPs) and their recycling after splicing require numerous assembly/recycling factors whose modes of action are often poorly understood. The intrinsically disordered TSSC4 protein has been identified as a nuclear-localized U5 snRNP and U4/U6-U5 tri-snRNP assembly/recycling factor, but how TSSC4's intrinsic disorder supports TSSC4 functions remains unknown. Using diverse interaction assays and cryogenic electron microscopy-based structural analysis, we show that TSSC4 employs four conserved, non-contiguous regions to bind the PRPF8 Jab1/MPN domain and the SNRNP200 helicase at functionally important sites. It thereby inhibits SNRNP200 helicase activity, spatially aligns the proteins, coordinates formation of a U5 sub-module and transiently blocks premature interaction of SNRNP200 with at least three other spliceosomal factors. Guided by the structure, we designed a TSSC4 variant that lacks stable binding to the PRPF8 Jab1/MPN domain or SNRNP200 in vitro. Comparative immunoprecipitation/mass spectrometry from HEK293 nuclear extract revealed distinct interaction profiles of wild type TSSC4 and the variant deficient in PRPF8/SNRNP200 binding with snRNP proteins, other spliceosomal proteins as well as snRNP assembly/recycling factors and chaperones. Our findings elucidate molecular strategies employed by an intrinsically disordered protein to promote snRNP assembly, and suggest multiple TSSC4-dependent stages during snRNP assembly/recycling. #1:  Journal: Acta Crystallogr D Struct Biol / Year: 2019 Journal: Acta Crystallogr D Struct Biol / Year: 2019Title: Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Authors: Liebschner D / Afonine PV / Baker ML / Bunkoczi G / Chen VB / Croll TI / Hintze B / Hung LW / Jain S / McCoy AJ / Moriarty NW / Oeffner RD / Poon BK / Prisant MG / Read RJ / Richardson JS / ...Authors: Liebschner D / Afonine PV / Baker ML / Bunkoczi G / Chen VB / Croll TI / Hintze B / Hung LW / Jain S / McCoy AJ / Moriarty NW / Oeffner RD / Poon BK / Prisant MG / Read RJ / Richardson JS / Richardson DC / Sammito MD / Sobolev OV / Stockwell DH / Terwilliger TC / Urzhumtsev AG / Videau LL / Williams CJ / Adams PD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13045.map.gz emd_13045.map.gz | 37.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13045-v30.xml emd-13045-v30.xml emd-13045.xml emd-13045.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13045_fsc.xml emd_13045_fsc.xml | 11.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13045.png emd_13045.png | 83.4 KB | ||
| Filedesc metadata |  emd-13045.cif.gz emd-13045.cif.gz | 7.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13045 http://ftp.pdbj.org/pub/emdb/structures/EMD-13045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13045 | HTTPS FTP |
-Related structure data
| Related structure data |  7os1MC  7os2C  7px3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13045.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13045.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Dimeric complex of Brr2 and a C-terminal fragment of Fbp21
| Entire | Name: Dimeric complex of Brr2 and a C-terminal fragment of Fbp21 |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric complex of Brr2 and a C-terminal fragment of Fbp21
| Supramolecule | Name: Dimeric complex of Brr2 and a C-terminal fragment of Fbp21 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: U5 small nuclear ribonucleoprotein 200 kDa helicase (Brr2)
| Supramolecule | Name: U5 small nuclear ribonucleoprotein 200 kDa helicase (Brr2) type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: WW domain-binding protein 4 (Fbp21)
| Supramolecule | Name: WW domain-binding protein 4 (Fbp21) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: U5 small nuclear ribonucleoprotein 200 kDa helicase
| Macromolecule | Name: U5 small nuclear ribonucleoprotein 200 kDa helicase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA helicase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 198.785797 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAEFDLDQGG EALAPRQVLD LEDLVFTQGS HFMANKRCQL PDGSFRRQRK GYEEVHVPAL KPKPFGSEEQ LLPVEKLPKY AQAGFEGFK TLNRIQSKLY RAALETDENL LLCAPTGAGK TNVALMCMLR EIGKHINMDG TINVDDFKII YIAPMRSLVQ E MVGSFGKR ...String: GAEFDLDQGG EALAPRQVLD LEDLVFTQGS HFMANKRCQL PDGSFRRQRK GYEEVHVPAL KPKPFGSEEQ LLPVEKLPKY AQAGFEGFK TLNRIQSKLY RAALETDENL LLCAPTGAGK TNVALMCMLR EIGKHINMDG TINVDDFKII YIAPMRSLVQ E MVGSFGKR LATYGITVAE LTGDHQLCKE EISATQIIVC TPEKWDIITR KGGERTYTQL VRLIILDEIH LLHDDRGPVL EA LVARAIR NIEMTQEDVR LIGLSATLPN YEDVATFLRV DPAKGLFYFD NSFRPVPLEQ TYVGITEKKA IKRFQIMNEI VYE KIMEHA GKNQVLVFVH SRKETGKTAR AIRDMCLEKD TLGLFLREGS ASTEVLRTEA EQCKNLELKD LLPYGFAIHH AGMT RVDRT LVEDLFADKH IQVLVSTATL AWGVNLPAHT VIIKGTQVYS PEKGRWTELG ALDILQMLGR AGRPQYDTKG EGILI TSHG ELQYYLSLLN QQLPIESQMV SKLPDMLNAE IVLGNVQNAK DAVNWLGYAY LYIRMLRSPT LYGISHDDLK GDPLLD QRR LDLVHTAALM LDKNNLVKYD KKTGNFQVTE LGRIASHYYI TNDTVQTYNQ LLKPTLSEIE LFRVFSLSSE FKNITVR EE EKLELQKLLE RVPIPVKESI EEPSAKINVL LQAFISQLKL EGFALMADMV YVTQSAGRLM RAIFEIVLNR GWAQLTDK T LNLCKMIDKR MWQSMCPLRQ FRKLPEEVVK KIEKKNFPFE RLYDLNHNEI GELIRMPKMG KTIHKYVHLF PKLELSVHL QPITRSTLKV ELTITPDFQW DEKVHGSSEA FWILVEDVDS EVILHHEYFL LKAKYAQDEH LITFFVPVFE PLPPQYFIRV VSDRWLSCE TQLPVSFRHL ILPEKYPPPT ELLDLQPLPV SALRNSAFES LYQDKFPFFN PIQTQVFNTV YNSDDNVFVG A PTGSGKTI CAEFAILRML LQSSEGRCVY ITPMEALAEQ VYMDWYEKFQ DRLNKKVVLL TGETSTDLKL LGKGNIIIST PE KWDILSR RWKQRKNVQN INLFVVDEVH LIGGENGPVL EVICSRMRYI SSQIERPIRI VALSSSLSNA KDVAHWLGCS ATS TFNFHP NVRPVPLELH IQGFNISHTQ TRLLSMAKPV YHAITKHSPK KPVIVFVPSR KQTRLTAIDI LTTCAADIQR QRFL HCTEK DLIPYLEKLS DSTLKETLLN GVGYLHEGLS PMERRLVEQL FSSGAIQVVV ASRSLCWGMN VAAHLVIIMD TQYYN GKIH AYVDYPIYDV LQMVGHANRP LQDDEGRCVI MCQGSKKDFF KKFLYEPLPV ESHLDHCMHD HFNAEIVTKT IENKQD AVD YLTWTFLYRR MTQNPNYYNL QGISHRHLSD HLSELVEQTL SDLEQSKCIS IEDEMDVAPL NLGMIAAYYY INYTTIE LF SMSLNAKTKV RGLIEIISNA AEYENIPIRH HEDNLLRQLA QKVPHKLNNP KFNDPHVKTN LLLQAHLSRM QLSAELQS D TEEILSKAIR LIQACVDVLS SNGWLSPALA AMELAQMVTQ AMWSKDSYLK QLPHFTSEHI KRCTDKGVES VFDIMEMED EERNALLQLT DSQIADVARF CNRYPNIELS YEVVDKDSIR SGGPVVVLVQ LEREEEVTGP VIAPLFPQKR EEGWWVVIGD AKSNSLISI KRLTLQQKAK VKLDFVAPAT GAHNYTLYFM SDAYMGCDQE YKFSVDVKEA UniProtKB: U5 small nuclear ribonucleoprotein 200 kDa helicase |
-Macromolecule #2: WW domain-binding protein 4
| Macromolecule | Name: WW domain-binding protein 4 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 20.208762 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMAFNPHTS DLPSSKVNEN SLGTLDESKS SDSHSDSDGE QEAEEGGVST ETEKPKIKFK EKNKNSDGGS DPETQKEKSI QKQNSLGSN EEKSKTLKKS NPYGEWQEIK QEVESHEEVD LELPSTENEY VSTSEADGGG EPKVVFKEKT VTSLGVMADG V APVFKKRR TENGKSRNLR QRGDDQ UniProtKB: WW domain-binding protein 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 7.6 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1877 / Average exposure time: 40.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 102 / Target criteria: CC, R.m.s.d. Bonds / Angles | ||||||
| Output model |  PDB-7os1: |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)