+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12887 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Folded elbow of cohesin | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cohesin / Elbow / Hinge / Smc1 / Smc3 / coiled coil / DNA / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationEstablishment of Sister Chromatid Cohesion / Resolution of Sister Chromatid Cohesion / DNA secondary structure binding / meiotic cohesin complex / establishment of meiotic sister chromatid cohesion / cohesin complex / mitotic cohesin complex / synaptonemal complex assembly / SUMOylation of DNA damage response and repair proteins / meiotic sister chromatid cohesion ...Establishment of Sister Chromatid Cohesion / Resolution of Sister Chromatid Cohesion / DNA secondary structure binding / meiotic cohesin complex / establishment of meiotic sister chromatid cohesion / cohesin complex / mitotic cohesin complex / synaptonemal complex assembly / SUMOylation of DNA damage response and repair proteins / meiotic sister chromatid cohesion / replication-born double-strand break repair via sister chromatid exchange / establishment of mitotic sister chromatid cohesion / reciprocal meiotic recombination / sister chromatid cohesion / mitotic sister chromatid cohesion / mitotic sister chromatid segregation / minor groove of adenine-thymine-rich DNA binding / G2/M transition of mitotic cell cycle / double-strand break repair / double-stranded DNA binding / cell division / protein kinase binding / ATP hydrolysis activity / DNA binding / ATP binding / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |   | |||||||||
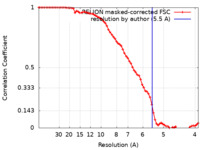
| Method | single particle reconstruction / cryo EM / Resolution: 5.5 Å | |||||||||
 Authors Authors | Gonzalez Llamazares A / Lee B-G / Collier J / Nasmyth KA / Lowe J | |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Folding of cohesin's coiled coil is important for Scc2/4-induced association with chromosomes. Authors: Naomi J Petela / Andres Gonzalez Llamazares / Sarah Dixon / Bin Hu / Byung-Gil Lee / Jean Metson / Heekyo Seo / Antonio Ferrer-Harding / Menelaos Voulgaris / Thomas Gligoris / James Collier ...Authors: Naomi J Petela / Andres Gonzalez Llamazares / Sarah Dixon / Bin Hu / Byung-Gil Lee / Jean Metson / Heekyo Seo / Antonio Ferrer-Harding / Menelaos Voulgaris / Thomas Gligoris / James Collier / Byung-Ha Oh / Jan Löwe / Kim A Nasmyth /   Abstract: Cohesin's association with and translocation along chromosomal DNAs depend on an ATP hydrolysis cycle driving the association and subsequent release of DNA. This involves DNA being 'clamped' by Scc2 ...Cohesin's association with and translocation along chromosomal DNAs depend on an ATP hydrolysis cycle driving the association and subsequent release of DNA. This involves DNA being 'clamped' by Scc2 and ATP-dependent engagement of cohesin's Smc1 and Smc3 head domains. Scc2's replacement by Pds5 abrogates cohesin's ATPase and has an important role in halting DNA loop extrusion. The ATPase domains of all SMC proteins are separated from their hinge dimerisation domains by 50-nm-long coiled coils, which have been observed to zip up along their entire length and fold around an elbow, thereby greatly shortening the distance between hinges and ATPase heads. Whether folding exists in vivo or has any physiological importance is not known. We present here a cryo-EM structure of the form of cohesin that reveals the structure of folded and zipped-up coils in unprecedented detail and shows that Scc2 can associate with Smc1's ATPase head even when it is fully disengaged from that of Smc3. Using cysteine-specific crosslinking, we show that cohesin's coiled coils are frequently folded in vivo, including when cohesin holds sister chromatids together. Moreover, we describe a mutation () within Smc1's hinge that alters how Scc2 and Pds5 interact with Smc1's hinge and that enables Scc2 to support loading in the absence of its normal partner Scc4. The mutant phenotype of loading without Scc4 is only explicable if loading depends on an association between Scc2/4 and cohesin's hinge, which in turn requires coiled coil folding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12887.map.gz emd_12887.map.gz | 2.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12887-v30.xml emd-12887-v30.xml emd-12887.xml emd-12887.xml | 13.3 KB 13.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12887_fsc.xml emd_12887_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_12887.png emd_12887.png | 23.7 KB | ||
| Filedesc metadata |  emd-12887.cif.gz emd-12887.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12887 http://ftp.pdbj.org/pub/emdb/structures/EMD-12887 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12887 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12887 | HTTPS FTP |
-Validation report
| Summary document |  emd_12887_validation.pdf.gz emd_12887_validation.pdf.gz | 346.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12887_full_validation.pdf.gz emd_12887_full_validation.pdf.gz | 346.3 KB | Display | |
| Data in XML |  emd_12887_validation.xml.gz emd_12887_validation.xml.gz | 9.6 KB | Display | |
| Data in CIF |  emd_12887_validation.cif.gz emd_12887_validation.cif.gz | 12.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12887 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12887 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12887 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12887 | HTTPS FTP |
-Related structure data
| Related structure data |  7ogtMC  7dg5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12887.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12887.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.96 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
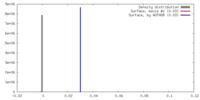
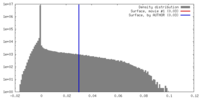
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cohesin
| Entire | Name: Cohesin |
|---|---|
| Components |
|
-Supramolecule #1: Cohesin
| Supramolecule | Name: Cohesin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Structural maintenance of chromosomes protein 1
| Macromolecule | Name: Structural maintenance of chromosomes protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 141.491781 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGRLVGLELS NFKSYRGVTK VGFGESNFTS IIGPNGSGKS NMMDAISFVL GVRSNHLRSN ILKDLIYRGV LNDENSDDYD NEGAASSNP QSAYVKAFYQ KGNKLVELMR IISRNGDTSY KIDGKTVSYK DYSIFLENEN ILIKAKNFLV FQGDVEQIAA Q SPVELSRM ...String: MGRLVGLELS NFKSYRGVTK VGFGESNFTS IIGPNGSGKS NMMDAISFVL GVRSNHLRSN ILKDLIYRGV LNDENSDDYD NEGAASSNP QSAYVKAFYQ KGNKLVELMR IISRNGDTSY KIDGKTVSYK DYSIFLENEN ILIKAKNFLV FQGDVEQIAA Q SPVELSRM FEEVSGSIQY KKEYEELKEK IEKLSKSATE SIKNRRRIHG ELKTYKEGIN KNEEYRKQLD KKNELQKFQA LW QLYHLEQ QKEELTDKLS ALNSEISSLK GKINNEMKSL QRSKSSFVKE SAVISKQKSK LDYIFKDKEK LVSDLRLIKV PQQ AAGKRI SHIEKRIESL QKDLQRQKTY VERFETQLKV VTRSKEAFEE EIKQSARNYD KFKLNENDLK TYNCLHEKYL TEGG SILEE KIAVLNNDKR EIQEELERFN KRADISKRRI TEELSITGEK LDTQLNDLRV SLNEKNALHT ERLHELKKLQ SDIES ANNQ EYDLNFKLRE TLVKIDDLSA NQRETMKERK LRENIAMLKR FFPGVKGLVH DLCHPKKEKY GLAVSTILGK NFDSVI VEN LTVAQECIAF LKKQRAGTAS FIPLDTIETE LPTLSLPDSQ DYILSINAID YEPEYEKAMQ YVCGDSIICN TLNIAKD LK WKKGIRGKLV TIEGALIHKA GLMTGGISGD ANNRWDKEEY QSLMSLKDKL LIQIDELSNG QRSNSIRARE VENSVSLL N SDIANLRTQV TQQKRSLDEN RLEIKYHNDL IEKEIQPKIT ELKKKLDDLE NTKDNLVKEK EALQNNIFKE FTSKIGFTI KEYENHSGEL MRQQSKELQQ LQKQILTVEN KLQFETDRLS TTQRRYEKAQ KDLENAQVEM KSLEEQEYAI EMKIGSIESK LEEHKNHLD ELQKKFVTKQ SELNSSEDIL EDMNSNLQVL KRERDGIKED IEKFDLERVT ALKNCKISNI NIPISSETTI D DLPISSTD NEAITISNSI DINYKGLPKK YKENNTDSAR KELEQKIHEV EEILNELQPN ARALERYDEA EGRFEVINNE TE QLKAEEK KILNQFLKIK KKRKELFEKT FDYVSDHLDA IYRELTKNPN SNVELAGGNA SLTIEDEDEP FNAGIKYHAT PPL KRFKDM EYLSGGEKTV AALALLFAIN SYQPSPFFVL DEVDAALDIT NVQRIAAYIR RHRNPDLQFI VISLKNTMFE KSDA LVGVY RQQQENSSKI ITLDLSNYAE UniProtKB: Structural maintenance of chromosomes protein 1 |
-Macromolecule #2: Structural maintenance of chromosomes protein 3
| Macromolecule | Name: Structural maintenance of chromosomes protein 3 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 141.539984 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYIKRVIIKG FKTYRNETII DNFSPHQNVI IGSNGSGKSN FFAAIRFVLS DDYSNLKREE RQGLIHQGSG GSVMSASVEI VFHDPDHSM ILPSGVLSRG DDEVTIRRTV GLKKDDYQLN DRNVTKGDIV RMLETAGFSM NNPYNIVPQG KIVALTNAKD K ERLQLLED ...String: MYIKRVIIKG FKTYRNETII DNFSPHQNVI IGSNGSGKSN FFAAIRFVLS DDYSNLKREE RQGLIHQGSG GSVMSASVEI VFHDPDHSM ILPSGVLSRG DDEVTIRRTV GLKKDDYQLN DRNVTKGDIV RMLETAGFSM NNPYNIVPQG KIVALTNAKD K ERLQLLED VVGAKSFEVK LKASLKKMEE TEQKKIQINK EMGELNSKLS EMEQERKELE KYNELERNRK IYQFTLYDRE LN EVINQME RLDGDYNNTV YSSEQYIQEL DKREDMIDQV SKKLSSIEAS LKIKNATDLQ QAKLRESEIS QKLTNVNVKI KDV QQQIES NEEQRNLDSA TLKEIKSIIE QRKQKLSKIL PRYQELTKEE AMYKLQLASL QQKQRDLILK KGEYARFKSK DERD TWIHS EIEELKSSIQ NLNELESQLQ MDRTSLRKQY SAIDEEIEEL IDSINGPDTK GQLEDFDSEL IHLKQKLSES LDTRK ELWR KEQKLQTVLE TLLSDVNQNQ RNVNETMSRS LANGIINVKE ITEKLKISPE SVFGTLGELI KVNDKYKTCA EVIGGN SLF HIVVDTEETA TLIMNELYRM KGGRVTFIPL NRLSLDSDVK FPSNTTTQIQ FTPLIKKIKY EPRFEKAVKH VFGKTIV VK DLGQGLKLAK KHKLNAITLD GDRADKRGVL TGGYLDQHKR TRLESLKNLN ESRSQHKKIL EELDFVRNEL NDIDTKID Q VNGNIRKVSN DRESVLTNIE VYRTSLNTKK NEKLILEESL NAIILKLEKL NTNRTFAQEK LNTFENDLLQ EFDSELSKE EKERLESLTK EISAAHNKLN ITSDALEGIT TTIDSLNAEL ESKLIPQEND LESKMSEVGD AFIFGLQDEL KELQLEKESV EKQHENAVL ELGTVQREIE SLIAEETNNK KLLEKANNQQ RLLLKKLDNF QKSVEKTMIK KTTLVTRREE LQQRIREIGL L PEDALVND FSDITSDQLL QRLNDMNTEI SGLKNVNKRA FENFKKFNER RKDLAERASE LDESKDSIQD LIVKLKQQKV NA VDSTFQK VSENFEAVFE RLVPRGTAKL IIHRKNDNAN DHDESIDVDM DAESNESQNG KDSEIMYTGV SISVSFNSKQ NEQ LHVEQL SGGQKTVCAI ALILAIQMVD PASFYLFDEI DAALDKQYRT AVATLLKELS KNAQFICTTF RTDMLQVADK FFRV KYENK ISTVIEVNRE EAIGFIRGSN KFAEV UniProtKB: Structural maintenance of chromosomes protein 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 42.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)