[English] 日本語
 Yorodumi
Yorodumi- EMDB-12716: Trimer of dimers TrwK/VirB4unbound C1 at 4.14A - Refinement witho... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Trimer of dimers TrwK/VirB4unbound C1 at 4.14A - Refinement without symmetry of TrwK/VirB4unbound trimer of dimers complex (with Hcp1) from the R388 type IV secretion system. | |||||||||||||||
 Map data Map data | TrwK/VirB4unbound trimer of dimer with HCP without symmetry sharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | type IV secretion system / type 4 secretion system / T4SS / inner membrane complex / inner membrane / R388 plasmid / conjugation / ATPase / bacterial secretion / secretion / secretion system / protein complex / VirB4 / TrwK / Hcp / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Salmonella dublin (bacteria) / Salmonella dublin (bacteria) /  Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||||||||
 Authors Authors | Vadakkepat AK / Mace K | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Cryo-EM structure of a type IV secretion system. Authors: Kévin Macé / Abhinav K Vadakkepat / Adam Redzej / Natalya Lukoyanova / Clasien Oomen / Nathalie Braun / Marta Ukleja / Fang Lu / Tiago R D Costa / Elena V Orlova / David Baker / Qian Cong ...Authors: Kévin Macé / Abhinav K Vadakkepat / Adam Redzej / Natalya Lukoyanova / Clasien Oomen / Nathalie Braun / Marta Ukleja / Fang Lu / Tiago R D Costa / Elena V Orlova / David Baker / Qian Cong / Gabriel Waksman /     Abstract: Bacterial conjugation is the fundamental process of unidirectional transfer of DNAs, often plasmid DNAs, from a donor cell to a recipient cell. It is the primary means by which antibiotic resistance ...Bacterial conjugation is the fundamental process of unidirectional transfer of DNAs, often plasmid DNAs, from a donor cell to a recipient cell. It is the primary means by which antibiotic resistance genes spread among bacterial populations. In Gram-negative bacteria, conjugation is mediated by a large transport apparatus-the conjugative type IV secretion system (T4SS)-produced by the donor cell and embedded in both its outer and inner membranes. The T4SS also elaborates a long extracellular filament-the conjugative pilus-that is essential for DNA transfer. Here we present a high-resolution cryo-electron microscopy (cryo-EM) structure of a 2.8 megadalton T4SS complex composed of 92 polypeptides representing 8 of the 10 essential T4SS components involved in pilus biogenesis. We added the two remaining components to the structural model using co-evolution analysis of protein interfaces, to enable the reconstitution of the entire system including the pilus. This structure describes the exceptionally large protein-protein interaction network required to assemble the many components that constitute a T4SS and provides insights on the unique mechanism by which they elaborate pili. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12716.map.gz emd_12716.map.gz | 23.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12716-v30.xml emd-12716-v30.xml emd-12716.xml emd-12716.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12716.png emd_12716.png | 95.4 KB | ||
| Filedesc metadata |  emd-12716.cif.gz emd-12716.cif.gz | 6.2 KB | ||
| Others |  emd_12716_additional_1.map.gz emd_12716_additional_1.map.gz emd_12716_half_map_1.map.gz emd_12716_half_map_1.map.gz emd_12716_half_map_2.map.gz emd_12716_half_map_2.map.gz | 22.6 MB 95.6 MB 95.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12716 http://ftp.pdbj.org/pub/emdb/structures/EMD-12716 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12716 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12716 | HTTPS FTP |
-Validation report
| Summary document |  emd_12716_validation.pdf.gz emd_12716_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12716_full_validation.pdf.gz emd_12716_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_12716_validation.xml.gz emd_12716_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_12716_validation.cif.gz emd_12716_validation.cif.gz | 13.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12716 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12716 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12716 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12716 | HTTPS FTP |
-Related structure data
| Related structure data |  7o42MC  7o3jC  7o3tC  7o3vC  7o41C  7o43C  7oiuC  7q1vC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12716.map.gz / Format: CCP4 / Size: 25 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12716.map.gz / Format: CCP4 / Size: 25 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TrwK/VirB4unbound trimer of dimer with HCP without symmetry sharpened map | ||||||||||||||||||||||||||||||||||||
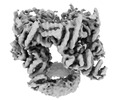
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||||||||||||||||||
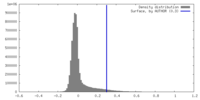
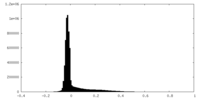
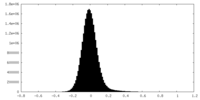
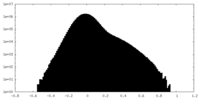
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: TrwK/VirB4unbound trimer of dimer with HCP without symmetry...
| File | emd_12716_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TrwK/VirB4unbound trimer of dimer with HCP without symmetry unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
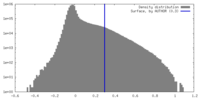
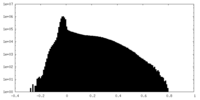
| Density Histograms |
-Half map: TrwK/VirB4unbound trimer of dimer - Half-A
| File | emd_12716_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TrwK/VirB4unbound trimer of dimer - Half-A | ||||||||||||
| Projections & Slices |
| ||||||||||||
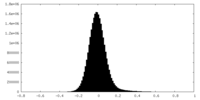
| Density Histograms |
-Half map: TrwK/VirB4unbound trimer of dimer - Half-A
| File | emd_12716_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | TrwK/VirB4unbound trimer of dimer - Half-A | ||||||||||||
| Projections & Slices |
| ||||||||||||
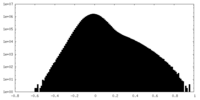
| Density Histograms |
- Sample components
Sample components
-Entire : Hexameric complex of Apo-TrwK/VirB4 with Hcp1
| Entire | Name: Hexameric complex of Apo-TrwK/VirB4 with Hcp1 |
|---|---|
| Components |
|
-Supramolecule #1: Hexameric complex of Apo-TrwK/VirB4 with Hcp1
| Supramolecule | Name: Hexameric complex of Apo-TrwK/VirB4 with Hcp1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Salmonella dublin (bacteria) Salmonella dublin (bacteria) |
-Macromolecule #1: TrwK protein,Protein hcp1
| Macromolecule | Name: TrwK protein,Protein hcp1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 |
| Molecular weight | Theoretical: 111.487844 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGAIESRKLL ASETPVGQFI PYSHHVTDTI ISTKNAEYLS VWKIDGRSHQ SASEADVFQW IRELNNTLRG ISSANLSLWT HIVRRRVYE YPDAEFDNVF CRQLDEKYRE SFTGYNLMVN DLYLTVVYRP VSDKVLSFFA KRERETPDQK KHRQESCIKA L EDINRTLG ...String: MGAIESRKLL ASETPVGQFI PYSHHVTDTI ISTKNAEYLS VWKIDGRSHQ SASEADVFQW IRELNNTLRG ISSANLSLWT HIVRRRVYE YPDAEFDNVF CRQLDEKYRE SFTGYNLMVN DLYLTVVYRP VSDKVLSFFA KRERETPDQK KHRQESCIKA L EDINRTLG QSFKRYGAEL LSVYEKGGHA FSAPLEFLAR LVNGEHIPMP ICRDRFSDYM AVNRPMFSKW GEVGELRSLT GL RRFGMLE IREYDDATEP GQLNVLLESD YEFVLTHSFS VLSRPAAKEY LQRHQKNLID ARDVATDQIE EIDEALNQLI SGH FVMGEH HCTLTVYGET VQQVRDNLAH ASAAMLDVAV LPKPVDLALE AGYWAQLPAN WQWRPRPAPI TSLNFLSFSP FHNF MSGKP TGNPWGPAVT ILKTVSGTPL YFNFHASKEE EDATDKRLLG NTMLIGQSSS GKTVLLGFLL AQAQKFKPTI VAFDK DRGM EISIRAMGGR YLPLKTGEPS GFNPFQLPPT HANLIFLKQF VKKLAAAGGE VTHRDEEEID QAITAMMSDS IDKSLR RLS LLLQFLPNPR SDDMDARPTV HARLVKWCEG GDYGWLFDNP TDALDLSTHQ IYGFDITEFL DNPEARTPVM MYLLYRT ES MIDGRRFMYV FDEFWKPLQD EYFEDLAKNK QKTIRKQNGI FVFATQEPSD ALESNIAKTL IQQCATYIFL ANPKADYE D YTQGFKLTDS EFELVRGLGE FSRRFLIKQG DQSALAEMNL GKFRTIVDGE TVERDFDDEL LVLSGTPDNA EIAESIIAE VGDDPAVWLP IFLDRVKAER SDVGSGSGSA VDMFIKIGDV KGESKDKTHA EEIDVLAWSW GMSQSGSMHM GGGGGAGKVN VQDLSFTKY IDKSTPNLMM ACSSGKHYPQ AKLTIRKAGG ENQVEYLIIT LKEVLVSSVS TGGSGGEDRL TENVTLNFAQ V QVDYQPQK ADGAKDGGPV KYGWNIRQNV QA UniProtKB: Type IV secretion system protein virB4, Protein hcp1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: CryoSPARC ab-initio |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 214048 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: OTHER / Details: Stochastic gradient descent (SGD) |
 Movie
Movie Controller
Controller












 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)