[English] 日本語
 Yorodumi
Yorodumi- EMDB-12584: CryoEM structure of the Nipah virus nucleocapsid spiral clam-shap... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12584 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the Nipah virus nucleocapsid spiral clam-shaped assembly | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Protein-RNA complex / Nucleocapsid / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative stranded viral RNA transcription / negative stranded viral RNA replication / helical viral capsid / viral nucleocapsid / molecular adaptor activity / host cell cytoplasm / ribonucleoprotein complex / structural molecule activity / RNA binding Similarity search - Function | |||||||||
| Biological species |  Nipah virus / Nipah virus /  | |||||||||
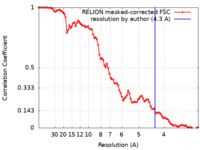
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Ker DS / Jenkins HT | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2021 Journal: PLoS Pathog / Year: 2021Title: CryoEM structure of the Nipah virus nucleocapsid assembly. Authors: De-Sheng Ker / Huw T Jenkins / Sandra J Greive / Alfred A Antson /  Abstract: Nipah and its close relative Hendra are highly pathogenic zoonotic viruses, storing their ssRNA genome in a helical nucleocapsid assembly formed by the N protein, a major viral immunogen. Here, we ...Nipah and its close relative Hendra are highly pathogenic zoonotic viruses, storing their ssRNA genome in a helical nucleocapsid assembly formed by the N protein, a major viral immunogen. Here, we report the first cryoEM structure for a Henipavirus RNA-bound nucleocapsid assembly, at 3.5 Å resolution. The helical assembly is stabilised by previously undefined N- and C-terminal segments, contributing to subunit-subunit interactions. RNA is wrapped around the nucleocapsid protein assembly with a periodicity of six nucleotides per protomer, in the "3-bases-in, 3-bases-out" conformation, with protein plasticity enabling non-sequence specific interactions. The structure reveals commonalities in RNA binding pockets and in the conformation of bound RNA, not only with members of the Paramyxoviridae family, but also with the evolutionarily distant Filoviridae Ebola virus. Significant structural differences with other Paramyxoviridae members are also observed, particularly in the position and length of the exposed α-helix, residues 123-139, which may serve as a valuable epitope for surveillance and diagnostics. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12584.map.gz emd_12584.map.gz | 6.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12584-v30.xml emd-12584-v30.xml emd-12584.xml emd-12584.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12584_fsc.xml emd_12584_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_12584.png emd_12584.png | 73.1 KB | ||
| Masks |  emd_12584_msk_1.map emd_12584_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12584.cif.gz emd-12584.cif.gz | 6.3 KB | ||
| Others |  emd_12584_half_map_1.map.gz emd_12584_half_map_1.map.gz emd_12584_half_map_2.map.gz emd_12584_half_map_2.map.gz | 27.5 MB 27.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12584 http://ftp.pdbj.org/pub/emdb/structures/EMD-12584 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12584 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12584 | HTTPS FTP |
-Related structure data
| Related structure data |  7nt6MC  7nt5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12584.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12584.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.572 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12584_msk_1.map emd_12584_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12584_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12584_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nipah virus nucleocapsid Protein-RNA complex
| Entire | Name: Nipah virus nucleocapsid Protein-RNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Nipah virus nucleocapsid Protein-RNA complex
| Supramolecule | Name: Nipah virus nucleocapsid Protein-RNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 1 MDa |
-Supramolecule #2: Nucleoprotein
| Supramolecule | Name: Nucleoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Nipah virus Nipah virus |
-Supramolecule #3: RNA (48-MER)
| Supramolecule | Name: RNA (48-MER) / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #4: RNA (42-MER)
| Supramolecule | Name: RNA (42-MER) / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Nucleoprotein
| Macromolecule | Name: Nucleoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 15 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nipah virus Nipah virus |
| Molecular weight | Theoretical: 60.609484 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLEVLFQG PAMSDIFEEA ASFRSYQSKL GRDGRASAAT ATLTTKIRIF VPATNSPELR WELTLFALDV IRSPSAAES MKVGAAFTLI SMYSERPGAL IRSLLNDPDI EAVIIDVGSM VNGIPVMERR GDKAQEEMEG LMRILKTARD S SKGKTPFV ...String: MGSSHHHHHH SSGLEVLFQG PAMSDIFEEA ASFRSYQSKL GRDGRASAAT ATLTTKIRIF VPATNSPELR WELTLFALDV IRSPSAAES MKVGAAFTLI SMYSERPGAL IRSLLNDPDI EAVIIDVGSM VNGIPVMERR GDKAQEEMEG LMRILKTARD S SKGKTPFV DSRAYGLRIT DMSTLVSAVI TIEAQIWILI AKAVTAPDTA EESETRRWAK YVQQKRVNPF FALTQQWLTE MR NLLSQSL SVRKFMVEIL IEVKKGGSAK GRAVEIISDI GNYVEETGMA GFFATIRFGL ETRYPALALN EFQSDLNTIK SLM LLYREI GPRAPYMVLL EESIQTKFAP GGYPLLWSFA MGVATTIDRS MGALNINRGY LEPMYFRLGQ KSARHHAGGI DQNM ANRLG LSSDQVAELA AAVQETSAGR QESNVQAREA KFAAGGVLIG GSDQDIDEGE EPIEQSGRQS VTFKREMSIS SLANS VPSS SVSTSGGTRL TNSLLNLRSR LAAKAAKEAA SSNATDDPAI SNRTQGESEK KNNQDLKPAQ NDLDFVRADV UniProtKB: Nucleoprotein |
-Macromolecule #2: RNA (48-MER)
| Macromolecule | Name: RNA (48-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 14.650994 KDa |
| Sequence | String: UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUU |
-Macromolecule #3: RNA (42-MER)
| Macromolecule | Name: RNA (42-MER) / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.814002 KDa |
| Sequence | String: UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UUUUUUUUUU UU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Average electron dose: 41.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.1 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)