+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12145 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of MsbA in Salipro with ADP vanadate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Lipid export MsbA / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMsbA transporter complex / lipopolysaccharide floppase activity / lipid translocation / ABC-type lipid A-core oligosaccharide transporter / lipopolysaccharide transport / ATPase-coupled lipid transmembrane transporter activity / ABC-type xenobiotic transporter activity / lipid transport / ATP-binding cassette (ABC) transporter complex / transmembrane transport ...MsbA transporter complex / lipopolysaccharide floppase activity / lipid translocation / ABC-type lipid A-core oligosaccharide transporter / lipopolysaccharide transport / ATPase-coupled lipid transmembrane transporter activity / ABC-type xenobiotic transporter activity / lipid transport / ATP-binding cassette (ABC) transporter complex / transmembrane transport / lipid binding / ATP hydrolysis activity / ATP binding / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
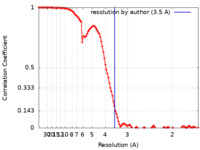
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Traore DAK / Tidow H | |||||||||
 Citation Citation |  Journal: FEBS J / Year: 2022 Journal: FEBS J / Year: 2022Title: Cryo-EM structure of MsbA in saposin-lipid nanoparticles (Salipro) provides insights into nucleotide coordination. Authors: Dominique-Maurice Kehlenbeck / Daouda A K Traore / Inokentijs Josts / Simon Sander / Martine Moulin / Michael Haertlein / Sylvain Prevost / V Trevor Forsyth / Henning Tidow /    Abstract: The ATP-binding cassette transporter MsbA is a lipid flippase, translocating lipid A, glycolipids, and lipopolysaccharides from the inner to the outer leaflet of the inner membrane of Gram-negative ...The ATP-binding cassette transporter MsbA is a lipid flippase, translocating lipid A, glycolipids, and lipopolysaccharides from the inner to the outer leaflet of the inner membrane of Gram-negative bacteria. It has been used as a model system for time-resolved structural studies as several MsbA structures in different states and reconstitution systems (detergent/nanodiscs/peptidiscs) are available. However, due to the limited resolution of the available structures, detailed structural information on the bound nucleotides has remained elusive. Here, we have reconstituted MsbA in saposin A-lipoprotein nanoparticles (Salipro) and determined the structure of ADP-vanadate-bound MsbA by single-particle cryo-electron microscopy to 3.5 Å resolution. This procedure has resulted in significantly improved resolution and enabled us to model all side chains and visualise detailed ADP-vanadate interactions in the nucleotide-binding domains. The approach may be applicable to other dynamic membrane proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12145.map.gz emd_12145.map.gz | 64.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12145-v30.xml emd-12145-v30.xml emd-12145.xml emd-12145.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12145_fsc.xml emd_12145_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_12145.png emd_12145.png | 40 KB | ||
| Filedesc metadata |  emd-12145.cif.gz emd-12145.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12145 http://ftp.pdbj.org/pub/emdb/structures/EMD-12145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12145 | HTTPS FTP |
-Related structure data
| Related structure data |  7bcwMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12145.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12145.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.827 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : MsbA in Salipro with ADP vanadate
| Entire | Name: MsbA in Salipro with ADP vanadate |
|---|---|
| Components |
|
-Supramolecule #1: MsbA in Salipro with ADP vanadate
| Supramolecule | Name: MsbA in Salipro with ADP vanadate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-dependent lipid A-core flippase
| Macromolecule | Name: ATP-dependent lipid A-core flippase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: ABC-type lipid A-core oligosaccharide transporter |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.486664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHEN LYFQGSMHND KDLSTWQTFR RLWPTIAPFK AGLIVAGVAL ILNAASDTFM LSLLKPLLDD GFGKTDRSVL VWMPLVVIG LMILRGITSY VSSYCISWVS GKVVMTMRRR LFGHMMGMPV SFFDKQSTGT LLSRITYDSE QVASSSSGAL I TVVREGAS ...String: MGHHHHHHEN LYFQGSMHND KDLSTWQTFR RLWPTIAPFK AGLIVAGVAL ILNAASDTFM LSLLKPLLDD GFGKTDRSVL VWMPLVVIG LMILRGITSY VSSYCISWVS GKVVMTMRRR LFGHMMGMPV SFFDKQSTGT LLSRITYDSE QVASSSSGAL I TVVREGAS IIGLFIMMFY YSWQLSIILI VLAPIVSIAI RVVSKRFRNI SKNMQNTMGQ VTTSAEQMLK GHKEVLIFGG QE VETKRFD KVSNRMRLQG MKMVSASSIS DPIIQLIASL ALAFVLYAAS FPSVMDSLTA GTITVVFSSM IALMRPLKSL TNV NAQFQR GMAACQTLFT ILDSEQEKDE GKRVIERATG DVEFRNVTFT YPGRDVPALR NINLKIPAGK TVALVGRSGS GKST IASLI TRFYDIDEGE ILMDGHDLRE YTLASLRNQV ALVSQNVHLF NDTVANNIAY ARTEQYSREQ IEEAARMAYA MDFIN KMDN GLDTVIGENG VLLSGGQRQR IAIARALLRD SPILILDEAT SALDTESERA IQAALDELQK NRTSLVIAHR LSTIEK ADE IVVVEDGVIV ERGTHNDLLE HRGVYAQLHK MQFGQ UniProtKB: ATP-dependent lipid A-core flippase |
-Macromolecule #2: VANADATE ION
| Macromolecule | Name: VANADATE ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: VO4 |
|---|---|
| Molecular weight | Theoretical: 114.939 Da |
| Chemical component information |  ChemComp-VN3: |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 5 / Number of copies: 2 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)