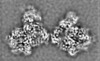
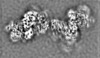
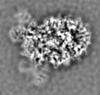
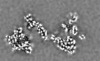
登録情報 データベース : EMDB / ID : EMD-11219タイトル Cryo-EM structure of DNA-PK dimer 細胞器官・細胞要素 : DNA-PK dimer細胞器官・細胞要素 : DNA-dependent protein kinase catalytic subunitタンパク質・ペプチド : DNA-dependent protein kinase catalytic subunit,DNA-PKcs細胞器官・細胞要素 : X-ray repair cross-complementing proteins 5 and 6タンパク質・ペプチド : X-ray repair cross-complementing protein 6タンパク質・ペプチド : X-ray repair cross-complementing protein 5細胞器官・細胞要素 : DNADNA : DNA (25-MER)DNA : DNA (27-MER)DNA : DNA (26-MER)DNA : DNA (28-MER) / / / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
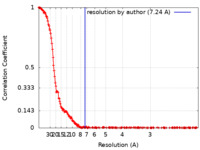
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)手法 / / 解像度 : 7.24 Å Chaplin AK / Hardwick SW 資金援助 Organization Grant number 国 Wellcome Trust 200814/Z/16/Z
ジャーナル : Nat Struct Mol Biol / 年 : 2021タイトル : Dimers of DNA-PK create a stage for DNA double-strand break repair.
著者 :
Amanda K Chaplin / Steven W Hardwick / Shikang Liang / Antonia Kefala Stavridi / Ales Hnizda / Lee R Cooper / Taiana Maia De Oliveira / Dimitri Y Chirgadze / Tom L Blundell / 要旨 :
DNA double-strand breaks are the most dangerous type of DNA damage and, if not repaired correctly, can lead to cancer. In humans, Ku70/80 recognizes DNA broken ends and recruits the DNA-dependent ... DNA double-strand breaks are the most dangerous type of DNA damage and, if not repaired correctly, can lead to cancer. In humans, Ku70/80 recognizes DNA broken ends and recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) to form DNA-dependent protein kinase holoenzyme (DNA-PK) in the process of non-homologous end joining (NHEJ). We present a 2.8-Å-resolution cryo-EM structure of DNA-PKcs, allowing precise amino acid sequence registration in regions uninterpreted in previous 4.3-Å X-ray maps. We also report a cryo-EM structure of DNA-PK at 3.5-Å resolution and reveal a dimer mediated by the Ku80 C terminus. Central to dimer formation is a domain swap of the conserved C-terminal helix of Ku80. Our results suggest a new mechanism for NHEJ utilizing a DNA-PK dimer to bring broken DNA ends together. Furthermore, drug inhibition of NHEJ in combination with chemo- and radiotherapy has proved successful, making these models central to structure-based drug targeting efforts. 履歴 登録 2020年6月23日 - ヘッダ(付随情報) 公開 2020年10月21日 - マップ公開 2020年10月21日 - 更新 2024年5月1日 - 現状 2024年5月1日 処理サイト : PDBe / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト) データ登録者
データ登録者 英国, 1件
英国, 1件  引用
引用 ジャーナル: Nat Struct Mol Biol / 年: 2021
ジャーナル: Nat Struct Mol Biol / 年: 2021
 ジャーナル: Nat.Struct.Mol.Biol. / 年: 2020
ジャーナル: Nat.Struct.Mol.Biol. / 年: 2020 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_11219.map.gz
emd_11219.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-11219-v30.xml
emd-11219-v30.xml emd-11219.xml
emd-11219.xml EMDBヘッダ
EMDBヘッダ emd_11219_fsc.xml
emd_11219_fsc.xml FSCデータファイル
FSCデータファイル emd_11219.png
emd_11219.png emd-11219.cif.gz
emd-11219.cif.gz emd_11219_additional_1.map.gz
emd_11219_additional_1.map.gz emd_11219_half_map_1.map.gz
emd_11219_half_map_1.map.gz emd_11219_half_map_2.map.gz
emd_11219_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-11219
http://ftp.pdbj.org/pub/emdb/structures/EMD-11219 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11219
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11219 emd_11219_validation.pdf.gz
emd_11219_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_11219_full_validation.pdf.gz
emd_11219_full_validation.pdf.gz emd_11219_validation.xml.gz
emd_11219_validation.xml.gz emd_11219_validation.cif.gz
emd_11219_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11219
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11219 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11219
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11219 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_11219.map.gz / 形式: CCP4 / 大きさ: 669.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_11219.map.gz / 形式: CCP4 / 大きさ: 669.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 ムービー
ムービー コントローラー
コントローラー

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)