+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11027 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Mrp antiporter complex | |||||||||
 Map data Map data | Composite map of the MRP dimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mrp antiporter / sodium/proton exchanger / bioenergetics / complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationsodium:proton antiporter activity / monoatomic cation transmembrane transporter activity / antiporter activity / sodium ion transport / NADH dehydrogenase (ubiquinone) activity / ATP synthesis coupled electron transport / proton transmembrane transport / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Anoxybacillus flavithermus WK1 (bacteria) / Anoxybacillus flavithermus WK1 (bacteria) /  Anoxybacillus flavithermus (strain DSM 21510 / WK1) (bacteria) Anoxybacillus flavithermus (strain DSM 21510 / WK1) (bacteria) | |||||||||
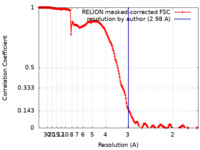
| Method | single particle reconstruction / cryo EM / Resolution: 2.98 Å | |||||||||
 Authors Authors | Steiner J / Sazanov LA | |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Structure and mechanism of the Mrp complex, an ancient cation/proton antiporter. Authors: Julia Steiner / Leonid Sazanov /  Abstract: Multiple resistance and pH adaptation (Mrp) antiporters are multi-subunit Na (or K)/H exchangers representing an ancestor of many essential redox-driven proton pumps, such as respiratory complex I. ...Multiple resistance and pH adaptation (Mrp) antiporters are multi-subunit Na (or K)/H exchangers representing an ancestor of many essential redox-driven proton pumps, such as respiratory complex I. The mechanism of coupling between ion or electron transfer and proton translocation in this large protein family is unknown. Here, we present the structure of the Mrp complex from solved by cryo-EM at 3.0 Å resolution. It is a dimer of seven-subunit protomers with 50 trans-membrane helices each. Surface charge distribution within each monomer is remarkably asymmetric, revealing probable proton and sodium translocation pathways. On the basis of the structure we propose a mechanism where the coupling between sodium and proton translocation is facilitated by a series of electrostatic interactions between a cation and key charged residues. This mechanism is likely to be applicable to the entire family of redox proton pumps, where electron transfer to substrates replaces cation movements. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11027.map.gz emd_11027.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11027-v30.xml emd-11027-v30.xml emd-11027.xml emd-11027.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11027_fsc.xml emd_11027_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_11027.png emd_11027.png | 202 KB | ||
| Filedesc metadata |  emd-11027.cif.gz emd-11027.cif.gz | 7.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11027 http://ftp.pdbj.org/pub/emdb/structures/EMD-11027 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11027 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11027 | HTTPS FTP |
-Related structure data
| Related structure data |  6z16MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11027.map.gz / Format: CCP4 / Size: 28 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11027.map.gz / Format: CCP4 / Size: 28 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map of the MRP dimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
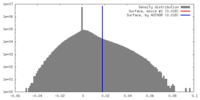
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Mrp dimer
+Supramolecule #1: Mrp dimer
+Macromolecule #1: Multisubunit Na+/H+ antiporter, A subunit
+Macromolecule #2: Multisubunit Na+/H+ antiporter, B subunit
+Macromolecule #3: Multisubunit Na+/H+ antiporter, C subunit
+Macromolecule #4: Multisubunit Na+/H+ antiporter, D subunit
+Macromolecule #5: Multisubunit Na+/H+ antiporter, E subunit
+Macromolecule #6: Multisubunit Na+/H+ antiporter, F subunit
+Macromolecule #7: Multisubunit Na+/H+ antiporter, G subunit
+Macromolecule #8: PHOSPHATIDYLETHANOLAMINE
+Macromolecule #9: POTASSIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.16 mg/mL |
|---|---|
| Buffer | pH: 6 |
| Grid | Model: Quantifoil R0.6/1 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 1.2 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 88.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)