+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10120 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of nucleotide-bound Tel1/ATM | |||||||||
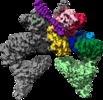
 Map data Map data | Tel1 Whole Dimer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinase / DNA Damage Response / CryoEM / Phosphatidylinositol-3-kinase-like kinase / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationDNA Damage/Telomere Stress Induced Senescence / Sensing of DNA Double Strand Breaks / Pexophagy / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / phosphatidylinositol-4-phosphate binding / telomeric DNA binding / signal transduction in response to DNA damage / telomere maintenance / DNA damage checkpoint signaling / double-strand break repair ...DNA Damage/Telomere Stress Induced Senescence / Sensing of DNA Double Strand Breaks / Pexophagy / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / phosphatidylinositol-4-phosphate binding / telomeric DNA binding / signal transduction in response to DNA damage / telomere maintenance / DNA damage checkpoint signaling / double-strand break repair / chromosome / chromatin organization / chromosome, telomeric region / protein kinase activity / non-specific serine/threonine protein kinase / protein serine kinase activity / DNA repair / protein serine/threonine kinase activity / DNA damage response / mitochondrion / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
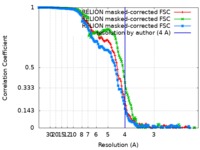
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Yates LA / Williams RM | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2020 Journal: Structure / Year: 2020Title: Cryo-EM Structure of Nucleotide-Bound Tel1 Unravels the Molecular Basis of Inhibition and Structural Rationale for Disease-Associated Mutations. Authors: Luke A Yates / Rhys M Williams / Sarem Hailemariam / Rafael Ayala / Peter Burgers / Xiaodong Zhang /   Abstract: Yeast Tel1 and its highly conserved human ortholog ataxia-telangiectasia mutated (ATM) are large protein kinases central to the maintenance of genome integrity. Mutations in ATM are found in ataxia- ...Yeast Tel1 and its highly conserved human ortholog ataxia-telangiectasia mutated (ATM) are large protein kinases central to the maintenance of genome integrity. Mutations in ATM are found in ataxia-telangiectasia (A-T) patients and ATM is one of the most frequently mutated genes in many cancers. Using cryoelectron microscopy, we present the structure of Tel1 in a nucleotide-bound state. Our structure reveals molecular details of key residues surrounding the nucleotide binding site and provides a structural and molecular basis for its intrinsically low basal activity. We show that the catalytic residues are in a productive conformation for catalysis, but the phosphatidylinositol 3-kinase-related kinase (PIKK) regulatory domain insert restricts peptide substrate access and the N-lobe is in an open conformation, thus explaining the requirement for Tel1 activation. Structural comparisons with other PIKKs suggest a conserved and common allosteric activation mechanism. Our work also provides a structural rationale for many mutations found in A-T and cancer. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10120.map.gz emd_10120.map.gz | 190.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10120-v30.xml emd-10120-v30.xml emd-10120.xml emd-10120.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10120_fsc_1.xml emd_10120_fsc_1.xml emd_10120_fsc_2.xml emd_10120_fsc_2.xml emd_10120_fsc_3.xml emd_10120_fsc_3.xml | 13.5 KB 13.5 KB 13.5 KB | Display Display Display |  FSC data file FSC data file |
| Images |  emd_10120.png emd_10120.png | 210.5 KB | ||
| Filedesc metadata |  emd-10120.cif.gz emd-10120.cif.gz | 6.5 KB | ||
| Others |  emd_10120_additional_1.map.gz emd_10120_additional_1.map.gz emd_10120_additional_2.map.gz emd_10120_additional_2.map.gz | 193.6 MB 193.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10120 http://ftp.pdbj.org/pub/emdb/structures/EMD-10120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10120 | HTTPS FTP |
-Related structure data
| Related structure data |  6s8fMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10120.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10120.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tel1 Whole Dimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.085 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Tel1 Kinase Core Dimer
| File | emd_10120_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tel1 Kinase Core Dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
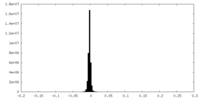
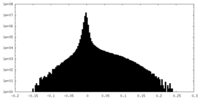
| Density Histograms |
-Additional map: Tel1 FATKIN Dimer
| File | emd_10120_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tel1 FATKIN Dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
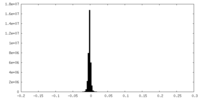
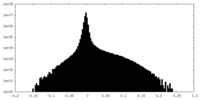
| Density Histograms |
- Sample components
Sample components
-Entire : Tel1/ATM
| Entire | Name: Tel1/ATM |
|---|---|
| Components |
|
-Supramolecule #1: Tel1/ATM
| Supramolecule | Name: Tel1/ATM / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Serine/threonine-protein kinase TEL1,Serine/threonine-protein kin...
| Macromolecule | Name: Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1,Serine/threonine-protein kinase TEL1 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 292.579406 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)GS IRGGKQRVFA TFIKCLQKLD SSNIINIMNS ISSYMAQV S YKNQSIIFYE IKSLFGPPQQ SIEKSAFYSL AMSMLSLVSY PSLVFSLEDM MTYSGFNHTR AFIQQALNKI TVAFRYQNL TELFEYCKFD LIMYWFNRTK VPTSKLEKEW DISLFGFADI HEFLGRYFVE ISAIYFSQGF NQKWILDMLH AITGNGDAYL VDNSYYLCI PLAFISGGVN ELIFDILPQI SGKTTVKYHK KYRLLMLKWI IRFTDLGSLT ELRSTVEKLF PTSYLSPYLF E NSSVSMRY QYPLHIPLAL GATLVQTQFA HEKNNTHEFK LLFLSVITDL EKTSTYIGKL RCARELKYLF VLYENVLVKS ST LNFIIIR LSKFLIDTQI HDEVITIFSS LLNLADKNTF EIEPSLPNLF CKIFIYLREN KQLSPSFQQA IKLLEHRDLI KIK TWKYCL DAIFGNIVQD DIYENTELLD ASDCGVDDVV LVSLLFSYAR RPVASKIGCS LSKAAAINIL KHHVPKEYLS KNFK LWFAA LSRRILQQEV QRERSTNFNN EVHLKNFEMV FRHPEQPHMI YQRISTFNKE AELYDSTEVF FISECILTYL VGYSI GNSE SEFCFRDNIM NENKDKVAPL DKDVLNAIYP LANNFGMESF ICDTYLSVNE PYNCWLSKFA RSLIHQISFN IPPIVC LYP LCKGSTAFCE LVLTDLFFLS TTYDPKSCLN WSNRIFTQIA MLLHVKDSEI KLKMLFNVIK MIRMGSRCKE RNCLRIY SS LDLQEICQIS LKIKEFKFGY LLFEEMNMPN IREMNINTLQ KIYECINDGD FLAGLPVPHS IEGVLNSINR IDSDTWKR F LFNNADFDAN YTTSLEEEKE SLIKATEDSG FYGLTSLLES RLSGSSDVYK WNLELGDWKL LTPKVVDSKA KGLYYAIKN LPQDVGFAEK SLEKSLLTIF DSRQHFISQT EWMDTLNAII EFIKIAAIPQ DVTSFPQTLM SIMKADKERL NTIDFYDHKT TLKSRHTLM NVLSRNSLDE NVKCSKYLRL GSIIQLANYV QLAIANGAPQ DALRNATLMS KTVKNIAKLY DDPSVVSQIE K LASFTSAN ALWESREYKA PVMIMRDLLA QNEKNISESI LYDDFKLLIN VPMDQIKARL VKWSSESRLE PAAAIYEKII VN WDINVED HESCSDVFYT LGSFLDEQAQ KLRSNGEIED REHRSYTGKS TLKALELIYK NTKLPENERK DAKRHYNRVL LQY NRDSEV LKALLLQKEK FLWHALHFYL NTLVFSNRYD NDIIDKFCGL WFENDDNSKI NQLLYKEIGT IPSWKFLPWV NQIA SKISM EENEFQKPLQ LTMKRLLYKL PYDSLYSVMS ILLYEKQSNK DTNISQKIQA VKKILLELQG YDRGAFAKKY LLPVQ EFCE MSVELANLKF VQNTKTLRLA NLKIGQYWLK QLNMEKLPLP TSNFTVKSSA DGRKARPYIV SVNETVGITT TGLSLP KIV TFNISDGTTQ KALMKGSNDD LRQDAIMEQV FQQVNKVLQN DKVLRNLDLG IRTYKVVPLG PKAGIIEFVA NSTSLHQ IL SKLHTNDKIT FDQARKGMKA VQTKSNEERL KAYLKITNEI KPQLRNFFFD SFPDPLDWFE AKKTYTKGVA ASSIVGYI L GLGDRHLNNI LLDCSTGEPI HIDLGIAFDQ GKLLPIPELV PFRLTRDIVD GFGVTGVDGL FRRSCERVYA VLRKDYVKV MCVLNILKWD PLYSWVMSPV KKYEHLFEEE HEITNFDNVS KFISNNDRNE NQESYRALKG VEEKLMGNGL SVESSVQDLI QQATDPSNL SVIYMGWSPF Y UniProtKB: Serine/threonine-protein kinase TEL1 |
-Macromolecule #2: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 2 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 88.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)