[English] 日本語
 Yorodumi
Yorodumi- EMDB-10089: Cryo-EM structure of Ctf18-1-8 in complex with the catalytic doma... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10089 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Ctf18-1-8 in complex with the catalytic domain of DNA polymerase epsilon (Class 2) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA polymerase / PCNA loader / protein complex / REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of mitotic sister chromatid cohesion / gene conversion / DNA replication initiation / epsilon DNA polymerase complex / telomere tethering at nuclear periphery / Ctf18 RFC-like complex / maintenance of DNA trinucleotide repeats / nucleotide-excision repair, DNA gap filling / SUMO binding / Activation of the pre-replicative complex ...maintenance of mitotic sister chromatid cohesion / gene conversion / DNA replication initiation / epsilon DNA polymerase complex / telomere tethering at nuclear periphery / Ctf18 RFC-like complex / maintenance of DNA trinucleotide repeats / nucleotide-excision repair, DNA gap filling / SUMO binding / Activation of the pre-replicative complex / DNA replication proofreading / single-stranded DNA 3'-5' DNA exonuclease activity / : / mitotic DNA replication checkpoint signaling / mitotic intra-S DNA damage checkpoint signaling / Hydrolases; Acting on ester bonds; Exodeoxyribonucleases producing 5'-phosphomonoesters / mitotic sister chromatid cohesion / leading strand elongation / nuclear replication fork / Dual incision in TC-NER / DNA replication initiation / chromosome, centromeric region / error-prone translesion synthesis / base-excision repair, gap-filling / replication fork / double-strand break repair via homologous recombination / base-excision repair / double-strand break repair via nonhomologous end joining / DNA-templated DNA replication / mitotic cell cycle / double-strand break repair / single-stranded DNA binding / 4 iron, 4 sulfur cluster binding / double-stranded DNA binding / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / nucleotide binding / mRNA binding / chromatin / ATP hydrolysis activity / mitochondrion / DNA binding / zinc ion binding / ATP binding / nucleus Similarity search - Function | |||||||||
| Biological species |   | |||||||||
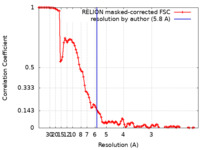
| Method | single particle reconstruction / cryo EM / Resolution: 5.8 Å | |||||||||
 Authors Authors | Grabarczyk DB / Song B | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2020 Journal: Nucleic Acids Res / Year: 2020Title: Ctf18-RFC and DNA Pol ϵ form a stable leading strand polymerase/clamp loader complex required for normal and perturbed DNA replication. Authors: Katy Stokes / Alicja Winczura / Boyuan Song / Giacomo De Piccoli / Daniel B Grabarczyk /   Abstract: The eukaryotic replisome must faithfully replicate DNA and cope with replication fork blocks and stalling, while simultaneously promoting sister chromatid cohesion. Ctf18-RFC is an alternative PCNA ...The eukaryotic replisome must faithfully replicate DNA and cope with replication fork blocks and stalling, while simultaneously promoting sister chromatid cohesion. Ctf18-RFC is an alternative PCNA loader that links all these processes together by an unknown mechanism. Here, we use integrative structural biology combined with yeast genetics and biochemistry to highlight the specific functions that Ctf18-RFC plays within the leading strand machinery via an interaction with the catalytic domain of DNA Pol ϵ. We show that a large and unusually flexible interface enables this interaction to occur constitutively throughout the cell cycle and regardless of whether forks are replicating or stalled. We reveal that, by being anchored to the leading strand polymerase, Ctf18-RFC can rapidly signal fork stalling to activate the S phase checkpoint. Moreover, we demonstrate that, independently of checkpoint signaling or chromosome cohesion, Ctf18-RFC functions in parallel to Chl1 and Mrc1 to protect replication forks and cell viability. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
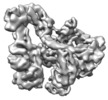
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10089.map.gz emd_10089.map.gz | 86.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10089-v30.xml emd-10089-v30.xml emd-10089.xml emd-10089.xml | 22.5 KB 22.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10089_fsc.xml emd_10089_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_10089.png emd_10089.png | 87.5 KB | ||
| Filedesc metadata |  emd-10089.cif.gz emd-10089.cif.gz | 7.1 KB | ||
| Others |  emd_10089_half_map_1.map.gz emd_10089_half_map_1.map.gz emd_10089_half_map_2.map.gz emd_10089_half_map_2.map.gz | 72.8 MB 72.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10089 http://ftp.pdbj.org/pub/emdb/structures/EMD-10089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10089 | HTTPS FTP |
-Related structure data
| Related structure data |  6s2fMC  6s1cC  6s2eC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10089.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10089.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0635 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
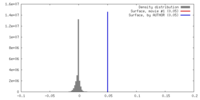
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_10089_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
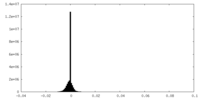
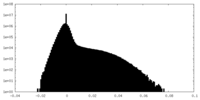
| Density Histograms |
-Half map: #2
| File | emd_10089_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
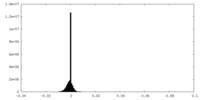
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of the catalytic domain of DNA polymerase epsilon with th...
| Entire | Name: Complex of the catalytic domain of DNA polymerase epsilon with the Ctf18-1-8 module of Ctf18-RFC |
|---|---|
| Components |
|
-Supramolecule #1: Complex of the catalytic domain of DNA polymerase epsilon with th...
| Supramolecule | Name: Complex of the catalytic domain of DNA polymerase epsilon with the Ctf18-1-8 module of Ctf18-RFC type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: DNA polymerase epsilon catalytic subunit A
| Macromolecule | Name: DNA polymerase epsilon catalytic subunit A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 140.39725 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKHHHHHHSA GLEVLFQGPG TGSEFELMMF GKKKNNGGSS TARYSAGNKY NTLSNNYALS AQQLLNASKI DDIDSMMGFE RYVPPQYNG RFDAKDIDQI PGRVGWLTNM HATLVSQETL SSGSNGGGNS NDGERVTTNQ GISGVDFYFL DEEGGSFKST V VYDPYFFI ...String: MKHHHHHHSA GLEVLFQGPG TGSEFELMMF GKKKNNGGSS TARYSAGNKY NTLSNNYALS AQQLLNASKI DDIDSMMGFE RYVPPQYNG RFDAKDIDQI PGRVGWLTNM HATLVSQETL SSGSNGGGNS NDGERVTTNQ GISGVDFYFL DEEGGSFKST V VYDPYFFI ACNDESRVND VEELVKKYLE SCLKSLQIIR KEDLTMDNHL LGLQKTLIKL SFVNSNQLFE ARKLLRPILQ DN ANNNVQR NIYNVAANGS EKVDAKHLIE DIREYDVPYH VRVSIDKDIR VGKWYKVTQQ GFIEDTRKIA FADPVVMAFA IAT TKPPLK FPDSAVDQIM MISYMIDGEG FLITNREIIS EDIEDFEYTP KPEYPGFFTI FNENDEVALL QRFFEHIRDV RPTV ISTFN GDFFDWPFIH NRSKIHGLDM FDEIGFAPDA EGEYKSSYCS HMDCFRWVKR DSYLPQGSQG LKAVTQSKLG YNPIE LDPE LMTPYAFEKP QHLSEYSVSD AVATYYLYMK YVHPFIFSLC TIIPLNPDET LRKGTGTLCE MLLMVQAYQH NILLPN KHT DPIERFYDGH LLESETYVGG HVESLEAGVF RSDLKNEFKI DPSAIDELLQ ELPEALKFSV EVENKSSVDK VTNFEEI KN QITQKLLELK ENNIRNELPL IYHVDVASMY PNIMTTNRLQ PDSIKAERDC ASCDFNRPGK TCARKLKWAW RGEFFPSK M DEYNMIKRAL QNETFPNKNK FSKKKVLTFD ELSYADQVIH IKKRLTEYSR KVYHRVKVSE IVEREAIVCQ RENPFYVDT VKSFRDRRYE FKGLAKTWKG NLSKIDPSDK HARDEAKKMI VLYDSLQLAH KVILNSFYGY VMRKGSRWYS MEMAGITCLT GATIIQMAR ALVERVGRPL ELDTDGIWCI LPKSFPETYF FTLENGKKLY LSYPCSMLNY RVHQKFTNHQ YQELKDPLNY I YETHSENT IFFEVDGPYK AMILPSSKEE GKGIKKRYAV FNEDGSLAEL KGFELKRRGE LQLIKNFQSD IFKVFLEGDT LE GCYSAVA SVCNRWLDVL DSHGLMLEDE DLVSLICENR SMSKTLKEYE GQKSTSITTA RRLGDFLGED MVKDKGLQCK YII SSKPFN APVTERAIPV AIFSADIPIK RSFLRRWTLD PSLEDLDIRT IIDWGYYRER LGSAIQKIIT IPAALQGVSN PVPR VEHPD WLKRKIATKE DKFK UniProtKB: DNA polymerase epsilon catalytic subunit A |
-Macromolecule #2: Chromosome transmission fidelity protein 8
| Macromolecule | Name: Chromosome transmission fidelity protein 8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 15.189688 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPSVDIDASQ WQKLTQSREK QTTVITPLGM MMLEIQGELE LPKDFASLAR RDSPNEGRFS EQDGETLIRF GSLQIDGERA TLFVGKKQR LLGKVTKLDV PMGIMHFNSK DNKVELVDVM KYKVIFKDRP LPIM UniProtKB: Chromosome transmission fidelity protein 8 |
-Macromolecule #3: Chromosome transmission fidelity protein 18
| Macromolecule | Name: Chromosome transmission fidelity protein 18 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 3.859329 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAMGNQTVKI WVKYNEGFSN AVRKNVTWNN LWE UniProtKB: Chromosome transmission fidelity protein 18 |
-Macromolecule #4: Sister chromatid cohesion protein DCC1
| Macromolecule | Name: Sister chromatid cohesion protein DCC1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 44.133785 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSINLHSAPE YDPSYKLIQL TPELLDIIQD PVQNHQLRFK SLDKDKSEVV LCSHDKTWVL KQRKHSNTVL LMREFVPEQP ITFDETLLF GLSKPYMDVV GFAKTESEFE TRETHGELNL NSVPIYNGEL DFSDKIMKRS STKVIGTLEE LLENSPCSAL E GISKWHKI ...String: MSINLHSAPE YDPSYKLIQL TPELLDIIQD PVQNHQLRFK SLDKDKSEVV LCSHDKTWVL KQRKHSNTVL LMREFVPEQP ITFDETLLF GLSKPYMDVV GFAKTESEFE TRETHGELNL NSVPIYNGEL DFSDKIMKRS STKVIGTLEE LLENSPCSAL E GISKWHKI GGSVKDGVLC ILSQDFLFKA LHVLLMSAMA ESLDLQHLNV EDTHHAVGKD IEDEFNPYTR EIIETVLNKF AV QEQEAEN NTWRLRIPFI AQWYGIQALR KYVSGISMPI DEFLIKWKSL FPPFFPCDID IDMLRGYHFK PTDKTVQYIA KST LPMDPK ERFKVLFRLQ SQWDLEDIKP LIEELNSRGM KIDSFIMKYA RRKRLGKKTV VTSR UniProtKB: Sister chromatid cohesion protein DCC1 |
-Macromolecule #5: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 5 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average exposure time: 75.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.4000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation coefficient | ||||||
| Output model |  PDB-6s2f: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)