[English] 日本語
 Yorodumi
Yorodumi- EMDB-0244: Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0244 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | |||||||||
 Map data Map data | cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA / polymerase D / Pyrococcus / REPLICATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationexodeoxyribonuclease I / DNA polymerase complex / intein-mediated protein splicing / single-stranded DNA 3'-5' DNA exonuclease activity / DNA catabolic process / DNA strand elongation involved in DNA replication / DNA-templated DNA replication / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA binding Similarity search - Function | |||||||||
| Biological species |   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.1 Å | |||||||||
 Authors Authors | Raia P / Carroni M | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2019 Journal: PLoS Biol / Year: 2019Title: Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases. Authors: Pierre Raia / Marta Carroni / Etienne Henry / Gérard Pehau-Arnaudet / Sébastien Brûlé / Pierre Béguin / Ghislaine Henneke / Erik Lindahl / Marc Delarue / Ludovic Sauguet /   Abstract: PolD is an archaeal replicative DNA polymerase (DNAP) made of a proofreading exonuclease subunit (DP1) and a larger polymerase catalytic subunit (DP2). Recently, we reported the individual crystal ...PolD is an archaeal replicative DNA polymerase (DNAP) made of a proofreading exonuclease subunit (DP1) and a larger polymerase catalytic subunit (DP2). Recently, we reported the individual crystal structures of the DP1 and DP2 catalytic cores, thereby revealing that PolD is an atypical DNAP that has all functional properties of a replicative DNAP but with the catalytic core of an RNA polymerase (RNAP). We now report the DNA-bound cryo-electron microscopy (cryo-EM) structure of the heterodimeric DP1-DP2 PolD complex from Pyrococcus abyssi, revealing a unique DNA-binding site. Comparison of PolD and RNAPs extends their structural similarities and brings to light the minimal catalytic core shared by all cellular transcriptases. Finally, elucidating the structure of the PolD DP1-DP2 interface, which is conserved in all eukaryotic replicative DNAPs, clarifies their evolutionary relationships with PolD and sheds light on the domain acquisition and exchange mechanism that occurred during the evolution of the eukaryotic replisome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0244.map.gz emd_0244.map.gz | 38.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0244-v30.xml emd-0244-v30.xml emd-0244.xml emd-0244.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0244.png emd_0244.png | 81.5 KB | ||
| Filedesc metadata |  emd-0244.cif.gz emd-0244.cif.gz | 7.7 KB | ||
| Others |  emd_0244_half_map_1.map.gz emd_0244_half_map_1.map.gz emd_0244_half_map_2.map.gz emd_0244_half_map_2.map.gz | 37.7 MB 37.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0244 http://ftp.pdbj.org/pub/emdb/structures/EMD-0244 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0244 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0244 | HTTPS FTP |
-Related structure data
| Related structure data |  6hmsMC  6hmfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0244.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0244.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
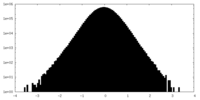
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
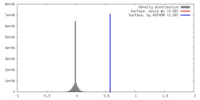
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: cryo-EM half map A of DNA polymerase D...
| File | emd_0244_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM half map A of DNA polymerase D from Pyrococcus abyssi in complex with DNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
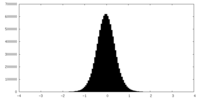
| Density Histograms |
-Half map: cryo-EM half map B of DNA polymerase D...
| File | emd_0244_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM half map B of DNA polymerase D from Pyrococcus abyssi in complex with DNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
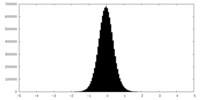
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between DNA polymerase D and DNA duplex
| Entire | Name: Complex between DNA polymerase D and DNA duplex |
|---|---|
| Components |
|
-Supramolecule #1: Complex between DNA polymerase D and DNA duplex
| Supramolecule | Name: Complex between DNA polymerase D and DNA duplex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|
-Supramolecule #2: DNA polymerase D
| Supramolecule | Name: DNA polymerase D / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) |
-Macromolecule #1: DNA polymerase II small subunit
| Macromolecule | Name: DNA polymerase II small subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) |
| Molecular weight | Theoretical: 45.078012 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: YEVKFVEAYA SLFKSRLSKL KRILRENPEI SNVVDIGKLN YVSGDEEVTI IGLVNSKRET NRGLIFEVED KTGIVKVFLP KDSEDYREA FKVLPDAVVA FKGFYSKKGI FFANKFYLPD VPLYRKQKPP LEEKVYAILI SDIHVGSREF CEKAFLKFLE W LNGHVESK ...String: YEVKFVEAYA SLFKSRLSKL KRILRENPEI SNVVDIGKLN YVSGDEEVTI IGLVNSKRET NRGLIFEVED KTGIVKVFLP KDSEDYREA FKVLPDAVVA FKGFYSKKGI FFANKFYLPD VPLYRKQKPP LEEKVYAILI SDIHVGSREF CEKAFLKFLE W LNGHVESK EEEEIVSRVK YLIIAGDVVD GIGIYPGQYS DLVIPDIFDQ YEALANLLAN VPEHITMFIG PGNADAARPA IP QPEFYKE YAKPIYKLKN AIIISNPAVI RLHGRDFLIA HGRGIEDVVS FVPGLTHHKP GLPMVELLKM RHLAPTFGGK VPI APDPED LLVIEEVPDL VQMGHVHVYD AVVYRGVQLV NSATWQAQTE FQKMVNIVPT PAKVPVVDVE SARVVKVLDF SGWC UniProtKB: DNA polymerase II small subunit |
-Macromolecule #2: DNA polymerase II large subunit,DNA polymerase II large subunit
| Macromolecule | Name: DNA polymerase II large subunit,DNA polymerase II large subunit type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) |
| Molecular weight | Theoretical: 144.418969 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MELPKEMEEY FEMLQREIDK AYEIAKKARA QGKDPSLDVE IPQATDMAGR VESLVGPPGV AKRIRELVKE YGKEIAALKI VDEIIEGKF GDLGSREKYA EQAVRTALAI LTEGIVSAPI EGIANVKIKR NTWADNSEYL ALYYAGPIRS SGGTAQALSV L VGDYVRRK ...String: MELPKEMEEY FEMLQREIDK AYEIAKKARA QGKDPSLDVE IPQATDMAGR VESLVGPPGV AKRIRELVKE YGKEIAALKI VDEIIEGKF GDLGSREKYA EQAVRTALAI LTEGIVSAPI EGIANVKIKR NTWADNSEYL ALYYAGPIRS SGGTAQALSV L VGDYVRRK LGLDRFKPSE KHIERMVEEV DLYHRAVTRL QYHPSPEEVR LAMRNIPIEI TGEATDDVEV SHRDVPGVET NQ LRGGAIL VLAEGVLQKA KKLVKYIDKM GIEGWEWLKE FVEAKEKGEP KEEGKEESLA ESTLEETKVE VDMGFYYSLY QKF KEEIAP SDKYAKEVIG GRPLFSDPSK PGGFRLRYGR SRASGFATWG INPATMILVD EFLAIGTQLK TERPGKGAVV TPVT TIEGP IVKLKDGSVL RVDDYNLALK VREDVEEILY LGDAVIAFGD FVENNQTLLP ANYCEEWWIL EFVKALKEIY EVHLE PFTE NEEESIEEAS DYLEIDPEFL KEMLRDPLRV KPPVELAIHF SEVLGIPLHP YYTLYWNSVE PKDVEKLWRL LKNYAE IEW SNFRGIKFAK KIVISQEKLG DSKRTLELLG LPHTVRDGNV IVDYPWAAAL LTPLGNLNWE FMAKPLYATI DIINENN EI KLRDRGISWI GARMGRPEKA KERKMKPPVQ VLFPIGLAGG SSRDIKKAAE EGKVAEVEIA FFKCPKCGHV GPEHLCPN C GTRKELLWVC PRCNAEYPES QAEGYNYTCP KCNVKLRPYA KRKIRPSELL NRAMENVKVY GVDKLKGVMG MTSGWKMPE PLEKGLLRAK NDVYVFKDGT IRFDATDAPI THFRPREIGV SVEKLRELGY THDFEGKPLV SEDQIVELKP QDIILSKEAG RYLLKVAKF VDDLLEKFYG LPRFYNAEKM EDLIGHLVIG LAPHTSAGIV GRIIGFVDAL VGYAHPYFHA AKRRNCDGDE D AVMLLLDA LLNFSRYYLP EKRGGKMDAP LVITTRLDPR EVDSEVHNMD IVRYYPLEFY EATYELKSPK ELVGVIERVE DR LGKPEMY YGLKFTHDTD DIALGPKMSL YKQLGDMEEK VRRQLEVAKR IRAVDEHGVA EKILNSHLIP DLRGNLRSFT RQE FRCVKC NTKFRRPPLN GKCPVCGGKI VLTVSKGAIE KYLGTAKMLV TEYNVKNYTR QRICLTERDI DSLFENVFPE TQLT LIVNP NDICQRLVMA RTGEVNKSGL LENLSNGSKK TEKAEKAEKP RKKSDEKPKK KRVISLEEFF SRKSK UniProtKB: DNA polymerase II large subunit, DNA polymerase II large subunit |
-Macromolecule #3: DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3')
| Macromolecule | Name: DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3') type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) |
| Molecular weight | Theoretical: 4.635998 KDa |
| Sequence | String: (DG)(DA)(DG)(DA)(DC)(DG)(DG)(DG)(DC)(DC) (DG)(DC)(DG)(DT)(DC) |
-Macromolecule #4: DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:   Pyrococcus abyssi (archaea) Pyrococcus abyssi (archaea) |
| Molecular weight | Theoretical: 4.851129 KDa |
| Sequence | String: (DT)(DG)(DA)(DC)(DG)(DC)(DG)(DG)(DC)(DC) (DC)(DG)(DT)(DC)(DT)(DC) |
-Macromolecule #5: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)