+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9851 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | membrane structure | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | alpha/beta translocator / membrane protein complex / Protein import / mitochondria / TRANSLOCASE | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial outer membrane translocase complex assembly / mitochondrial outer membrane translocase complex / protein insertion into mitochondrial outer membrane / protein transmembrane transport / : / porin activity / pore complex / protein import into mitochondrial matrix / protein transmembrane transporter activity / monoatomic ion transport ...mitochondrial outer membrane translocase complex assembly / mitochondrial outer membrane translocase complex / protein insertion into mitochondrial outer membrane / protein transmembrane transport / : / porin activity / pore complex / protein import into mitochondrial matrix / protein transmembrane transporter activity / monoatomic ion transport / mitochondrial intermembrane space / mitochondrial outer membrane / mitochondrion / cytosol Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.81 Å | ||||||||||||||||||
 Authors Authors | Araiso Y / Tsutsumi A | ||||||||||||||||||
| Funding support |  Japan, 5 items Japan, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structure of the mitochondrial import gate reveals distinct preprotein paths. Authors: Yuhei Araiso / Akihisa Tsutsumi / Jian Qiu / Kenichiro Imai / Takuya Shiota / Jiyao Song / Caroline Lindau / Lena-Sophie Wenz / Haruka Sakaue / Kaori Yunoki / Shin Kawano / Junko Suzuki / ...Authors: Yuhei Araiso / Akihisa Tsutsumi / Jian Qiu / Kenichiro Imai / Takuya Shiota / Jiyao Song / Caroline Lindau / Lena-Sophie Wenz / Haruka Sakaue / Kaori Yunoki / Shin Kawano / Junko Suzuki / Marilena Wischnewski / Conny Schütze / Hirotaka Ariyama / Toshio Ando / Thomas Becker / Trevor Lithgow / Nils Wiedemann / Nikolaus Pfanner / Masahide Kikkawa / Toshiya Endo /      Abstract: The translocase of the outer mitochondrial membrane (TOM) is the main entry gate for proteins. Here we use cryo-electron microscopy to report the structure of the yeast TOM core complex at 3.8-Å ...The translocase of the outer mitochondrial membrane (TOM) is the main entry gate for proteins. Here we use cryo-electron microscopy to report the structure of the yeast TOM core complex at 3.8-Å resolution. The structure reveals the high-resolution architecture of the translocator consisting of two Tom40 β-barrel channels and α-helical transmembrane subunits, providing insight into critical features that are conserved in all eukaryotes. Each Tom40 β-barrel is surrounded by small TOM subunits, and tethered by two Tom22 subunits and one phospholipid. The N-terminal extension of Tom40 forms a helix inside the channel; mutational analysis reveals its dual role in early and late steps in the biogenesis of intermembrane-space proteins in cooperation with Tom5. Each Tom40 channel possesses two precursor exit sites. Tom22, Tom40 and Tom7 guide presequence-containing preproteins to the exit in the middle of the dimer, whereas Tom5 and the Tom40 N extension guide preproteins lacking a presequence to the exit at the periphery of the dimer. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9851.map.gz emd_9851.map.gz | 16.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9851-v30.xml emd-9851-v30.xml emd-9851.xml emd-9851.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9851_fsc.xml emd_9851_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_9851.png emd_9851.png | 114.7 KB | ||
| Masks |  emd_9851_msk_1.map emd_9851_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-9851.cif.gz emd-9851.cif.gz | 6.5 KB | ||
| Others |  emd_9851_half_map_1.map.gz emd_9851_half_map_1.map.gz emd_9851_half_map_2.map.gz emd_9851_half_map_2.map.gz | 19.7 MB 19.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9851 http://ftp.pdbj.org/pub/emdb/structures/EMD-9851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9851 | HTTPS FTP |
-Related structure data
| Related structure data |  6jnfMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10332 (Title: Cryo-EM structure of the translocator of the outer mitochondrial membrane EMPIAR-10332 (Title: Cryo-EM structure of the translocator of the outer mitochondrial membraneData size: 1.9 TB Data #1: Unaligned multi-frame micrographs of yeast TOM complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9851.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9851.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.2915 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
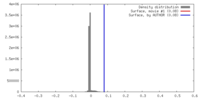
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9851_msk_1.map emd_9851_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
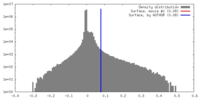
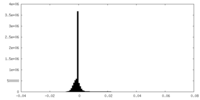
| Density Histograms |
-Half map: #1
| File | emd_9851_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
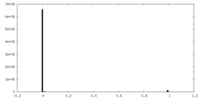
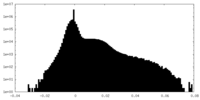
| Density Histograms |
-Half map: #2
| File | emd_9851_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
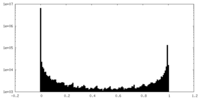
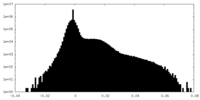
| Density Histograms |
- Sample components
Sample components
-Entire : TOM complex
| Entire | Name: TOM complex |
|---|---|
| Components |
|
-Supramolecule #1: TOM complex
| Supramolecule | Name: TOM complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Mitochondrial import receptor subunit TOM40
| Macromolecule | Name: Mitochondrial import receptor subunit TOM40 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.071141 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAPTPLAEA SQIPTIPALS PLTAKQSKGN FFSSNPISSF VVDTYKQLHS HRQSLELVNP GTVENLNKEV SRDVFLSQYF FTGLRADLN KAFSMNPAFQ TSHTFSIGSQ ALPKYAFSAL FANDNLFAQG NIDNDLSVSG RLNYGWDKKN ISKVNLQISD G QPTMCQLE ...String: MSAPTPLAEA SQIPTIPALS PLTAKQSKGN FFSSNPISSF VVDTYKQLHS HRQSLELVNP GTVENLNKEV SRDVFLSQYF FTGLRADLN KAFSMNPAFQ TSHTFSIGSQ ALPKYAFSAL FANDNLFAQG NIDNDLSVSG RLNYGWDKKN ISKVNLQISD G QPTMCQLE QDYQASDFSV NVKTLNPSFS EKGEFTGVAV ASFLQSVTPQ LALGLETLYS RTDGSAPGDA GVSYLTRYVS KK QDWIFSG QLQANGALIA SLWRKVAQNV EAGIETTLQA GMVPITDPLM GTPIGIQPTV EGSTTIGAKY EYRQSVYRGT LDS NGKVAC FLERKVLPTL SVLFCGEIDH FKNDTKIGCG LQFETAGNQE LLMLQQGLDA DGNPLQALPQ L UniProtKB: Mitochondrial import receptor subunit TOM40 |
-Macromolecule #2: Mitochondrial import receptor subunit TOM7
| Macromolecule | Name: Mitochondrial import receptor subunit TOM7 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 6.876955 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSFLPSFILS DESKERISKI LTLTHNVAHY GWIPFVLYLG WAHTSNRPNF LNLLSPLPSV UniProtKB: Mitochondrial import receptor subunit TOM7 |
-Macromolecule #3: Mitochondrial import receptor subunit TOM22
| Macromolecule | Name: Mitochondrial import receptor subunit TOM22 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 18.481139 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVELTEIKDD VVQLDEPQFS RNQAIVEEKA SATNNDVVDD EDDSDSDFED EFDENETLLD RIVALKDIVP PGKRQTISNF FGFTSSFVR NAFTKSGNLA WTLTTTALLL GVPLSLSILA EQQLIEMEKT FDLQSDANNI LAQGEKDAAA TANGSPGHHH H HHHHHH UniProtKB: Mitochondrial import receptor subunit TOM22 |
-Macromolecule #4: Mitochondrial import receptor subunit TOM5
| Macromolecule | Name: Mitochondrial import receptor subunit TOM5 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 5.993924 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFGLPQQEVS EEEKRAHQEQ TEKTLKQAAY VAAFLWVSPM IWHLVKKQWK UniProtKB: Mitochondrial import receptor subunit TOM5 |
-Macromolecule #5: Mitochondrial import receptor subunit TOM6
| Macromolecule | Name: Mitochondrial import receptor subunit TOM6 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 6.41046 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDGMFAMPGA AAGAASPQQP KSRFQAFKES PLYTIALNGA FFVAGVAFIQ SPLMDMLAPQ L UniProtKB: Mitochondrial import receptor subunit TOM6 |
-Macromolecule #6: (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradeca...
| Macromolecule | Name: (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate type: ligand / ID: 6 / Number of copies: 1 / Formula: 46E |
|---|---|
| Molecular weight | Theoretical: 635.853 Da |
| Chemical component information |  ChemComp-46E: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5.4 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 279 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)