[English] 日本語
 Yorodumi
Yorodumi- EMDB-9116: CryoEM reconstruction of native lens connexin-46/50 at 3.4 angstr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9116 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM reconstruction of native lens connexin-46/50 at 3.4 angstrom resolution | |||||||||
 Map data Map data | Cx46/50 D6 symmetrized map, 3.4 angstrom resolution, sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ion channel / gap junction / cell communication / connexin / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationgap junction-mediated intercellular transport / gap junction hemi-channel activity / connexin complex / gap junction channel activity / visual perception / cell-cell signaling / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
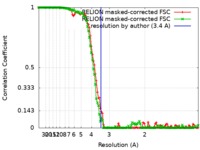
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Myers JB / Reichow SL | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2018 Journal: Nature / Year: 2018Title: Structure of native lens connexin 46/50 intercellular channels by cryo-EM. Authors: Janette B Myers / Bassam G Haddad / Susan E O'Neill / Dror S Chorev / Craig C Yoshioka / Carol V Robinson / Daniel M Zuckerman / Steve L Reichow /   Abstract: Gap junctions establish direct pathways for cell-to-cell communication through the assembly of twelve connexin subunits that form intercellular channels connecting neighbouring cells. Co-assembly of ...Gap junctions establish direct pathways for cell-to-cell communication through the assembly of twelve connexin subunits that form intercellular channels connecting neighbouring cells. Co-assembly of different connexin isoforms produces channels with unique properties and enables communication across cell types. Here we used single-particle cryo-electron microscopy to investigate the structural basis of connexin co-assembly in native lens gap junction channels composed of connexin 46 and connexin 50 (Cx46/50). We provide the first comparative analysis to connexin 26 (Cx26), which-together with computational studies-elucidates key energetic features governing gap junction permselectivity. Cx46/50 adopts an open-state conformation that is distinct from the Cx26 crystal structure, yet it appears to be stabilized by a conserved set of hydrophobic anchoring residues. 'Hot spots' of genetic mutations linked to hereditary cataract formation map to the core structural-functional elements identified in Cx46/50, suggesting explanations for many of the disease-causing effects. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9116.map.gz emd_9116.map.gz | 13.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9116-v30.xml emd-9116-v30.xml emd-9116.xml emd-9116.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9116_fsc_1.xml emd_9116_fsc_1.xml emd_9116_fsc_2.xml emd_9116_fsc_2.xml | 11 KB 10.9 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_9116.png emd_9116.png | 139.2 KB | ||
| Masks |  emd_9116_msk_1.map emd_9116_msk_1.map emd_9116_msk_2.map emd_9116_msk_2.map | 125 MB 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-9116.cif.gz emd-9116.cif.gz | 6.1 KB | ||
| Others |  emd_9116_additional_1.map.gz emd_9116_additional_1.map.gz emd_9116_additional_2.map.gz emd_9116_additional_2.map.gz emd_9116_additional_3.map.gz emd_9116_additional_3.map.gz | 20.1 MB 95.5 MB 95.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9116 http://ftp.pdbj.org/pub/emdb/structures/EMD-9116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9116 | HTTPS FTP |
-Related structure data
| Related structure data |  6mhqMC  6mhyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10212 (Title: CryoEM reconstruction of native lens connexin-46/50 at 3.4 angstrom resolution EMPIAR-10212 (Title: CryoEM reconstruction of native lens connexin-46/50 at 3.4 angstrom resolutionData size: 774.5 Data #1: Unaligned frame stacks - MP38 dataset 01 [micrographs - multiframe] Data #2: Unaligned frame stacks - MP38 dataset 02 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9116.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9116.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cx46/50 D6 symmetrized map, 3.4 angstrom resolution, sharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.665 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
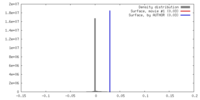
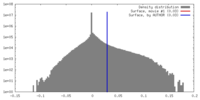
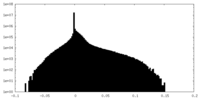
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9116_msk_1.map emd_9116_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
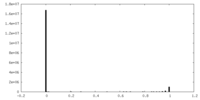
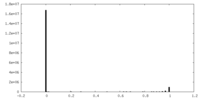
| Density Histograms |
-Mask #2
| File |  emd_9116_msk_2.map emd_9116_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
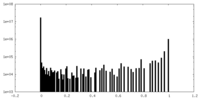
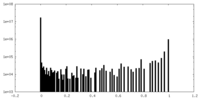
| Density Histograms |
-Additional map: Cx46/50 D6 symmetrized map, 3.5 angstrom resolution, sharpened
| File | emd_9116_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cx46/50 D6 symmetrized map, 3.5 angstrom resolution, sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Cx46/50 D6 symmetrized map, 3.5 angstrom resolution, unprocessed
| File | emd_9116_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cx46/50 D6 symmetrized map, 3.5 angstrom resolution, unprocessed | ||||||||||||
| Projections & Slices |
| ||||||||||||
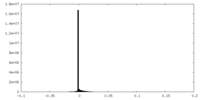
| Density Histograms |
-Additional map: Cx46/50 D6 symmetrized map, 3.4 angstrom resolution, unprocessed
| File | emd_9116_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cx46/50 D6 symmetrized map, 3.4 angstrom resolution, unprocessed | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Connexin-46 gap junction
| Entire | Name: Connexin-46 gap junction |
|---|---|
| Components |
|
-Supramolecule #1: Connexin-46 gap junction
| Supramolecule | Name: Connexin-46 gap junction / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Location in cell: C-terminal truncated version isolated from lens core |
| Molecular weight | Theoretical: 450 KDa |
-Macromolecule #1: Gap junction alpha-3 protein, connexin-46
| Macromolecule | Name: Gap junction alpha-3 protein, connexin-46 / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.968516 KDa |
| Sequence | String: MGDWSFLGRL LENAQEHSTV IGKVWLTVLF IFRILVLGAA AEEVWGDEQS DFTCNTQQPG CENVCYDRAF PISHVRFWVL QIIFVSTPT LIYLGHVLHL VRMEEKRKER EEEPPKAAGP AEEHQDPAPV RDDRGKVRIA GALLRTYVFN IIFKTLFEVG F IAGQYFLY ...String: MGDWSFLGRL LENAQEHSTV IGKVWLTVLF IFRILVLGAA AEEVWGDEQS DFTCNTQQPG CENVCYDRAF PISHVRFWVL QIIFVSTPT LIYLGHVLHL VRMEEKRKER EEEPPKAAGP AEEHQDPAPV RDDRGKVRIA GALLRTYVFN IIFKTLFEVG F IAGQYFLY GFQLKPLYRC DRWPCPNTVD CFISRPTEKT IFILFMLAVA CVSLLLNVLE IYHLGWKKLK QGMTSPFRPD TP GSRAGSA KPMGGSPLLL PPNSAPPAVT IGFPPYYAPS ASSLGQASAP GYPEPPLPAA LPGTPGTPGT PGTLGGGGGN QGL RAPAQN CANREAEPQT SARKASPPAS TP UniProtKB: Gap junction alpha-3 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.35 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: 10 sec wait before blotting, 4.0 second blot before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 30 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Average exposure time: 10.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.25 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL |
| Output model |  PDB-6mhq:  PDB-6mhy: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)