[English] 日本語
 Yorodumi
Yorodumi- EMDB-7550: Rabbit muscle aldolase at 2.4A resolution (17dec27a 205k particle... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7550 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rabbit muscle aldolase at 2.4A resolution (17dec27a 205k particles, all images) | |||||||||
 Map data Map data | rabbit muscle aldolase at 2.4 A resolution (17dec27a <25nm ice thickness) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
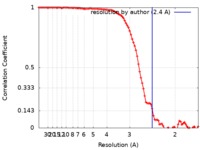
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Kim LK / Rice WJ / Eng ET / Kopylov M / Cheng A / Raczkowski AM / Jordan KD / Bobe D / Potter CS / Carragher B | |||||||||
 Citation Citation |  Journal: Front Mol Biosci / Year: 2018 Journal: Front Mol Biosci / Year: 2018Title: Benchmarking cryo-EM Single Particle Analysis Workflow. Authors: Laura Y Kim / William J Rice / Edward T Eng / Mykhailo Kopylov / Anchi Cheng / Ashleigh M Raczkowski / Kelsey D Jordan / Daija Bobe / Clinton S Potter / Bridget Carragher /  Abstract: Cryo electron microscopy facilities running multiple instruments and serving users with varying skill levels need a robust and reliable method for benchmarking both the hardware and software ...Cryo electron microscopy facilities running multiple instruments and serving users with varying skill levels need a robust and reliable method for benchmarking both the hardware and software components of their single particle analysis workflow. The workflow is complex, with many bottlenecks existing at the specimen preparation, data collection and image analysis steps; the samples and grid preparation can be of unpredictable quality, there are many different protocols for microscope and camera settings, and there is a myriad of software programs for analysis that can depend on dozens of settings chosen by the user. For this reason, we believe it is important to benchmark the entire workflow, using a standard sample and standard operating procedures, on a regular basis. This provides confidence that all aspects of the pipeline are capable of producing maps to high resolution. Here we describe benchmarking procedures using a test sample, rabbit muscle aldolase. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7550.map.gz emd_7550.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7550-v30.xml emd-7550-v30.xml emd-7550.xml emd-7550.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7550_fsc.xml emd_7550_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_7550.png emd_7550.png | 43.9 KB | ||
| Others |  emd_7550_half_map_1.map.gz emd_7550_half_map_1.map.gz emd_7550_half_map_2.map.gz emd_7550_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7550 http://ftp.pdbj.org/pub/emdb/structures/EMD-7550 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7550 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7550 | HTTPS FTP |
-Validation report
| Summary document |  emd_7550_validation.pdf.gz emd_7550_validation.pdf.gz | 78.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7550_full_validation.pdf.gz emd_7550_full_validation.pdf.gz | 78 KB | Display | |
| Data in XML |  emd_7550_validation.xml.gz emd_7550_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7550 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7550 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7550 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7550 | HTTPS FTP |
-Related structure data
| Related structure data |  7528C  7541C  7551C  7562C  7614C  7615C  7616C  7617C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10184 (Title: Benchmarking cryo-EM single particle analysis workflow EMPIAR-10184 (Title: Benchmarking cryo-EM single particle analysis workflowData size: 85.6 Data #1: aligned micrographs of rabbit muscle aldolase from 17dec27a [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7550.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7550.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | rabbit muscle aldolase at 2.4 A resolution (17dec27a <25nm ice thickness) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
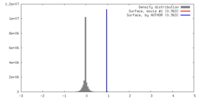
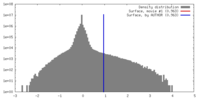
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: rabbit muscle aldolase at 2.4 A resolution (17dec27a...
| File | emd_7550_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | rabbit muscle aldolase at 2.4 A resolution (17dec27a <25nm ice thickness), half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
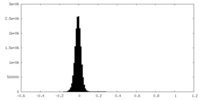
| Density Histograms |
-Half map: rabbit muscle aldolase at 2.4 A resolution (17dec27a...
| File | emd_7550_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | rabbit muscle aldolase at 2.4 A resolution (17dec27a <25nm ice thickness), half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
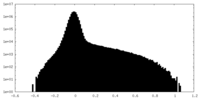
| Density Histograms |
- Sample components
Sample components
-Entire : Rabbit muscle aldolase
| Entire | Name: Rabbit muscle aldolase |
|---|---|
| Components |
|
-Supramolecule #1: Rabbit muscle aldolase
| Supramolecule | Name: Rabbit muscle aldolase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 150 KDa |
-Macromolecule #1: rabbit muscle aldolase
| Macromolecule | Name: rabbit muscle aldolase / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO / EC number: fructose-bisphosphate aldolase |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: PHSHPALTPE QKKELSDIAH RIVAPGKGIL AADESTGSIA KRLQSIGTEN TEENRRFYRQ LLLTADDRVN PCIGGVILFH ETLYQKADD GRPFPQVIKS KGGVVGIKVD KGVVPLAGTN GETTTQGLDG LSERCAQYKK DGADFAAWRC VLKIGEHTPS A LAIMENAN ...String: PHSHPALTPE QKKELSDIAH RIVAPGKGIL AADESTGSIA KRLQSIGTEN TEENRRFYRQ LLLTADDRVN PCIGGVILFH ETLYQKADD GRPFPQVIKS KGGVVGIKVD KGVVPLAGTN GETTTQGLDG LSERCAQYKK DGADFAAWRC VLKIGEHTPS A LAIMENAN VLARYASICQ QNGIVPIVEP EILPDGDHDL KRCQYVTEKV LAAVYKALSD HHIYLEGTLL KPNMVTPGHA CT QKYSHEE IAMATVTALR RTVPPAVTGV TFLSGGQSEE EASINLNAIN KCPLLKPWAL TFSYGRALQA SALKAWGGKK ENL KAAQEE YVKRALANSL ACQGKYTPSG QAGAAASESL FISNHAY |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)