[English] 日本語
 Yorodumi
Yorodumi- EMDB-6950: Subtomogram average of a nucleosome with long linker DNA from a V... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6950 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of a nucleosome with long linker DNA from a Volta phase contrast cryotomogram of a cryo-FIB lamella of an interphase HeLa cell | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 21.0 Å | |||||||||||||||
 Authors Authors | Cai S / Bock D / Pilhofer M / Gan L | |||||||||||||||
| Funding support |  Singapore, Singapore,  Switzerland, 4 items Switzerland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Mol Biol Cell / Year: 2018 Journal: Mol Biol Cell / Year: 2018Title: The in situ structures of mono-, di-, and trinucleosomes in human heterochromatin. Authors: Shujun Cai / Désirée Böck / Martin Pilhofer / Lu Gan /   Abstract: The in situ three-dimensional organization of chromatin at the nucleosome and oligonucleosome levels is unknown. Here we use cryo-electron tomography to determine the in situ structures of HeLa ...The in situ three-dimensional organization of chromatin at the nucleosome and oligonucleosome levels is unknown. Here we use cryo-electron tomography to determine the in situ structures of HeLa nucleosomes, which have canonical core structures and asymmetric, flexible linker DNA. Subtomogram remapping suggests that sequential nucleosomes in heterochromatin follow irregular paths at the oligonucleosome level. This basic principle of higher-order repressive chromatin folding is compatible with the conformational variability of the two linker DNAs at the single-nucleosome level. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6950.map.gz emd_6950.map.gz | 69.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6950-v30.xml emd-6950-v30.xml emd-6950.xml emd-6950.xml | 11.9 KB 11.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6950.png emd_6950.png | 53.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6950 http://ftp.pdbj.org/pub/emdb/structures/EMD-6950 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6950 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6950 | HTTPS FTP |
-Related structure data
| Related structure data |  6948C  6949C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10179 (Title: The in situ structures of mono-, di-, and trinucleosomes in human heterochromatin EMPIAR-10179 (Title: The in situ structures of mono-, di-, and trinucleosomes in human heterochromatinData size: 1.9 Data #1: Tilt series of a cryo-FIB lamella of an interphase HeLa cell [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6950.map.gz / Format: CCP4 / Size: 85.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6950.map.gz / Format: CCP4 / Size: 85.9 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 6.9 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HeLa cell, interphase
| Entire | Name: HeLa cell, interphase |
|---|---|
| Components |
|
-Supramolecule #1: HeLa cell, interphase
| Supramolecule | Name: HeLa cell, interphase / type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Strain: CCL2- / Organ: cervix Homo sapiens (human) / Strain: CCL2- / Organ: cervix |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Details: yeast-extract supplemented |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Sampling interval: 5.0 µm / Number grids imaged: 1 / Number real images: 61 / Average electron dose: 2.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 14493 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus min: 0.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


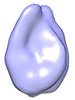








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















