+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vibrio cholerae DdmD apo complex | |||||||||
 Map data Map data | Unsharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Helicase / Nuclease / Complex / Effector / IMMUNE SYSTEM | |||||||||
| Function / homology | Helicase/UvrB N-terminal domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
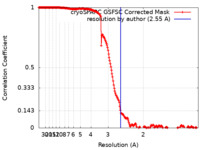
| Method | single particle reconstruction / cryo EM / Resolution: 2.55 Å | |||||||||
 Authors Authors | Loeff L / Jinek M | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Molecular mechanism of plasmid elimination by the DdmDE defense system. Authors: Luuk Loeff / David W Adams / Christelle Chanez / Sandrine Stutzmann / Laurie Righi / Melanie Blokesch / Martin Jinek /  Abstract: Seventh-pandemic strains contain two pathogenicity islands that encode the DNA defense modules DdmABC and DdmDE. In this study, we used cryogenic electron microscopy to determine the mechanistic ...Seventh-pandemic strains contain two pathogenicity islands that encode the DNA defense modules DdmABC and DdmDE. In this study, we used cryogenic electron microscopy to determine the mechanistic basis for plasmid defense by DdmDE. The helicase-nuclease DdmD adopts an autoinhibited dimeric architecture. The prokaryotic Argonaute protein DdmE uses a DNA guide to target plasmid DNA. The structure of the DdmDE complex, validated by in vivo mutational studies, shows that DNA binding by DdmE triggers disassembly of the DdmD dimer and loading of monomeric DdmD onto the nontarget DNA strand. In vitro studies indicate that DdmD translocates in the 5'-to-3' direction, while partially degrading the plasmid DNA. These findings provide critical insights into the mechanism of DdmDE systems in plasmid elimination. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50090.map.gz emd_50090.map.gz | 361.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50090-v30.xml emd-50090-v30.xml emd-50090.xml emd-50090.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_50090_fsc.xml emd_50090_fsc.xml | 19 KB | Display |  FSC data file FSC data file |
| Images |  emd_50090.png emd_50090.png | 165.5 KB | ||
| Masks |  emd_50090_msk_1.map emd_50090_msk_1.map | 729 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50090.cif.gz emd-50090.cif.gz | 6.5 KB | ||
| Others |  emd_50090_additional_1.map.gz emd_50090_additional_1.map.gz emd_50090_half_map_1.map.gz emd_50090_half_map_1.map.gz emd_50090_half_map_2.map.gz emd_50090_half_map_2.map.gz | 684.8 MB 675.9 MB 675.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50090 http://ftp.pdbj.org/pub/emdb/structures/EMD-50090 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50090 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50090 | HTTPS FTP |
-Related structure data
| Related structure data |  9ezxMC  9ezyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_50090.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50090.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
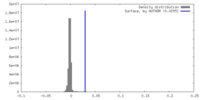
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50090_msk_1.map emd_50090_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
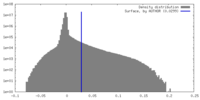
| Density Histograms |
-Additional map: Sharpened EM map
| File | emd_50090_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened EM map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_50090_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_50090_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dimeric complex of the DdmD protein
| Entire | Name: Dimeric complex of the DdmD protein |
|---|---|
| Components |
|
-Supramolecule #1: Dimeric complex of the DdmD protein
| Supramolecule | Name: Dimeric complex of the DdmD protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 266 KDa |
-Macromolecule #1: Helicase/UvrB N-terminal domain-containing protein
| Macromolecule | Name: Helicase/UvrB N-terminal domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 136.427594 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMNVSIEE FTHFDFQLVP EPSPLDLVIT EPLKNHIEVN GVKSGALLPL PFQTGIGKTY TALNFLLQQM LEQVRSELKE ENTGKKSKR LLYYVTDSVD NVVSAKADLL KLIEKQTVKG EPRFTLEQQE YLKAQIVHLP NQSEQLLQCS DAVLNDVLIG F NLNAERDV ...String: SNAMNVSIEE FTHFDFQLVP EPSPLDLVIT EPLKNHIEVN GVKSGALLPL PFQTGIGKTY TALNFLLQQM LEQVRSELKE ENTGKKSKR LLYYVTDSVD NVVSAKADLL KLIEKQTVKG EPRFTLEQQE YLKAQIVHLP NQSEQLLQCS DAVLNDVLIG F NLNAERDV QAEWSAISGL RRHASNPEVK ISLNRQAGYF YRNLIDRLQK KQKGADRVLL SGSLLASVET LLPGEKIRNG SA HVAFLTT SKFLKGFHNT RSRYSPLRDL SGAVLIIDEI DKQNQVILSE LCKQQAQDLI WAIRTLRANF RDHQLESSPR YDK IEDLFE PLRERLEEFG TNWNLAFAFN TEGANLNERP VRLFSDRSFT HVSSATHKLS LKSDFLRRKN LIFSDEKVEG SLIE KHGLL TRFVNEADVI YQWFLGTMRK AVFQYWENVR GLEIEVRENR SLEGTFQEAV QSLLTHFNLQ EFESAVYESF DTRGL RQSA GGKANKLSSS KSYHHTGLKL VEVAHNQGTR DTVNCKASFL NTSPSGVLAD MVDAGAVILG ISATARADTV IHNFDF KYL NERLGNKLLS LSREQKQRVN NYYHSRRNYK DNGVVLTVKY LNSRDAFLDA LLEEYKPEAR SSHFILNHYL GIAESEQ AF VRSWLSKLLA SIKAFISSPD NRYMLSLLNR TLDTTRQNIN DFIQFCCDKW AKEFNVKTKT FFGVNADWMR LVGYDEIS K HLNTELGKVV VFSTYASMGA GKNPDYAVNL ALEGESLISV ADVTYSTQLR SDIDSIYLEK PTQLLLSDDY SHTANQLCQ FHQILSLQEN GELSPKSAEN WCRQQLMGMS RERSLQQYHQ TSDYQSAVRK YIEQAVGRAG RTSLKRKQIL LFVDSGLKEI LAEESRDPS LFSHEYVALV NKAKSAGKSI VEDRAVRRLF NLAQRNNKDG MLSIKALVHR LHNQPASKSD IQEWQDIRTQ L LRYPTVAF QPERFNRLYL QSMTKGYYRY QGNLDGDPNS FEFFDRVPYG DMVSEEDCSL ATLVQNQYVR PWFERKGFAC SW QKEANVM TPIMFTNIYK GALGEQAVEA VLTAFDFTFE EVPNSIYERF DNRVIFAGIE QPIWLDSKYW KHEGNESSEG YSS KIALVE EEFGPSKFIY VNALGDTSKP IRYLNSCFVE TSPQLAKVIE IPALIDDSNA DTNRTAVQEL IKWLHHS UniProtKB: Helicase/UvrB N-terminal domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV | |||||||||
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 59.98 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 130000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model / Details: Initial model was generated with alpha fold |
|---|---|
| Output model |  PDB-9ezx: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)