+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4989 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
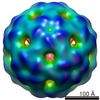
| Title | T=3 MS2 Virus-like particle | |||||||||
 Map data Map data | MS2 bacteriophage T=3 virus-like particle map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MS2 / T=3 / BACTERIOPHAGE / VLP / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of viral translation / T=3 icosahedral viral capsid / regulation of translation / structural molecule activity / RNA binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Escherichia phage MS2 (virus) / Escherichia phage MS2 (virus) /  Escherichia virus MS2 Escherichia virus MS2 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | de Martin Garrido N / Ramlaul K | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Microbiol / Year: 2020 Journal: Mol Microbiol / Year: 2020Title: Bacteriophage MS2 displays unreported capsid variability assembling T = 4 and mixed capsids. Authors: Natàlia de Martín Garrido / Michael A Crone / Kailash Ramlaul / Paul A Simpson / Paul S Freemont / Christopher H S Aylett /  Abstract: Bacteriophage MS2 is a positive-sense, single-stranded RNA virus encapsulated in an asymmetric T = 3 pseudo-icosahedral capsid. It infects Escherichia coli through the F-pilus, in which it binds ...Bacteriophage MS2 is a positive-sense, single-stranded RNA virus encapsulated in an asymmetric T = 3 pseudo-icosahedral capsid. It infects Escherichia coli through the F-pilus, in which it binds through a maturation protein incorporated into its capsid. Cryogenic electron microscopy has previously shown that its genome is highly ordered within virions, and that it regulates the assembly process of the capsid. In this study, we have assembled recombinant MS2 capsids with non-genomic RNA containing the capsid incorporation sequence, and investigated the structures formed, revealing that T = 3, T = 4 and mixed capsids between these two triangulation numbers are generated, and resolving structures of T = 3 and T = 4 capsids to 4 Å and 6 Å respectively. We conclude that the basic MS2 capsid can form a mix of T = 3 and T = 4 structures, supporting a role for the ordered genome in favouring the formation of functional T = 3 virions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4989.map.gz emd_4989.map.gz | 198 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4989-v30.xml emd-4989-v30.xml emd-4989.xml emd-4989.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_4989.png emd_4989.png | 74.7 KB | ||
| Filedesc metadata |  emd-4989.cif.gz emd-4989.cif.gz | 5.9 KB | ||
| Others |  emd_4989_half_map_1.map.gz emd_4989_half_map_1.map.gz emd_4989_half_map_2.map.gz emd_4989_half_map_2.map.gz | 168.9 MB 168.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4989 http://ftp.pdbj.org/pub/emdb/structures/EMD-4989 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4989 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4989 | HTTPS FTP |
-Related structure data
| Related structure data |  6rrsMC  4990C  6rrtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10706 (Title: Mixed capsids of MS2 virions / Data size: 113.1 EMPIAR-10706 (Title: Mixed capsids of MS2 virions / Data size: 113.1 Data #1: MS2 virion-like particles [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4989.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4989.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MS2 bacteriophage T=3 virus-like particle map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0277 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: MS2 bacteriophage T=3 virus-like particle unfiltered half-map 1
| File | emd_4989_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MS2 bacteriophage T=3 virus-like particle unfiltered half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: MS2 bacteriophage T=3 virus-like particle unfiltered half-map 2
| File | emd_4989_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | MS2 bacteriophage T=3 virus-like particle unfiltered half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia virus MS2
| Entire | Name:  Escherichia virus MS2 Escherichia virus MS2 |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia virus MS2
| Supramolecule | Name: Escherichia virus MS2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 329852 / Sci species name: Escherichia virus MS2 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|
-Macromolecule #1: Capsid protein
| Macromolecule | Name: Capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage MS2 (virus) Escherichia phage MS2 (virus) |
| Molecular weight | Theoretical: 13.869659 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNFTQFVL VDNGGTGDVT VAPSNFANGV AEWISSNSRS QAYKVTCSVR QSSAQNRKYT IKVEVPKVAT QTVGGVELPV AAWRSYLNM ELTIPIFATN SDCELIVKAM QGLLKDGNPI PSAIAANSGI Y UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 / Details: 10 mM Tris-HCl pH 7.5, 1 mM EDTA, 100 mM NaCl |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI FALCON I (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 4 / Number real images: 1040 / Average exposure time: 4.0 sec. / Average electron dose: 80.0 e/Å2 Details: Frames were obtained with a beam-splitter and recorded at 100kx |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: -2.0 µm / Nominal defocus min: -0.3 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)







































